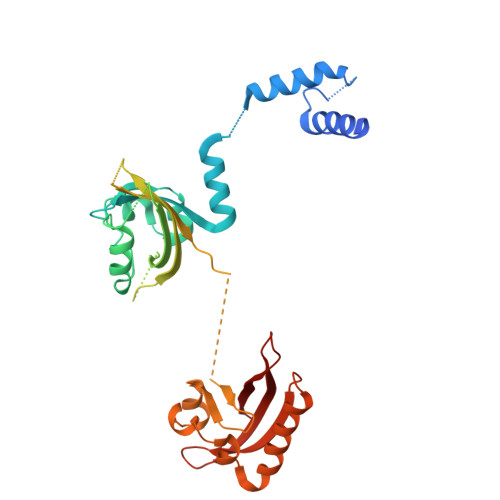
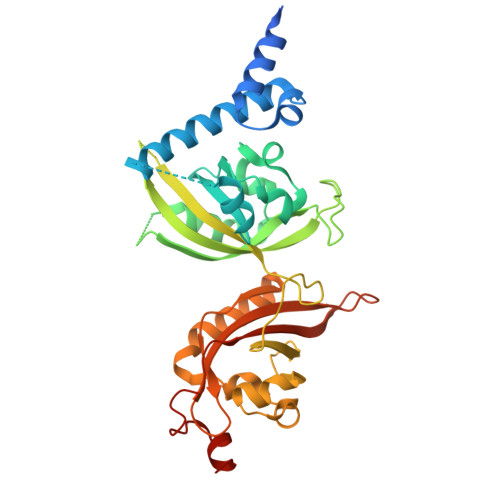
Structure-Based Optimization of HIF-2 alpha Agonists That Synergistically Enhance Erythropoietin Production with PHD Inhibitors.
Guo, M., Xu, W., Bai, Z., Chen, K., Li, F., Jin, Z., Chen, X., Shang, Q., Zhang, M., Diao, X., Xu, D., Zhang, H., Mu, K., Wang, Y., Ly, T., Li, L., Zhu, Y., Song, C., Zhao, H., Wan, X., Zhang, Y., Zhuang, J., Wu, D.(2025) J Med Chem 68: 21346-21365
- PubMed: 41047546
- DOI: https://doi.org/10.1021/acs.jmedchem.5c01257
- Primary Citation of Related Structures:
9LJX, 9LJY, 9LOH, 9LOI - PubMed Abstract:
Hypoxia-inducible factor 2α (HIF-2α) is a crucial transcription factor regulating various physiological processes, including angiogenesis and erythropoiesis. The activity of HIF-2α is mainly controlled through oxygen-dependent protein hydroxylation, mediated by prolyl hydroxylase domain (PHD) enzymes, leading to subsequent HIF-2α ubiquitination and degradation. While several small-molecule PHD inhibitors have already been clinically applied in renal anemia treatment by indirectly activating the HIF-2α pathway, direct HIF-2α agonists remain largely unexplored. Here, we developed derivatives of HIF-2α agonist M1001, identifying SD-10 through molecular/cellular evaluations. By determining its cocrystal structure with the heterodimeric HIF-2 protein complex, we precisely characterized its molecular mechanism of action. Notably, SD-10 exhibited remarkable synergy with Daprodustat, an approved PHD inhibitor, in stimulating erythropoietin (EPO) secretion both in cellular models and animal studies. These findings not only provide insights into HIF-2α activation mechanism, but also offer a promising lead compound for developing innovative treatments for renal anemia.
- State Key Laboratory of Microbial Technology, Microbial Technology Institute, Shandong University, Qingdao 266237, China.
Organizational Affiliation: