A monoclonal antibody targeting conserved regions of pre-fusion protein cross-neutralizes Nipah and Hendra virus variants.
Li, T., Xu, H., Zhang, M., Nie, J., Liao, B., Xie, J., Jiang, Y., Liu, Y., Ge, P., Zhao, C., Sun, Z., Bai, Y., Tang, M., Su, X., Wang, Y., Huang, W.(2025) Antiviral Res 240: 106215-106215
- PubMed: 40541691
- DOI: https://doi.org/10.1016/j.antiviral.2025.106215
- Primary Citation of Related Structures:
9LNG - PubMed Abstract:
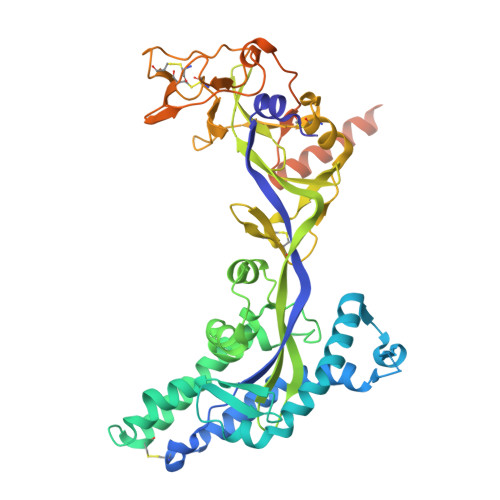
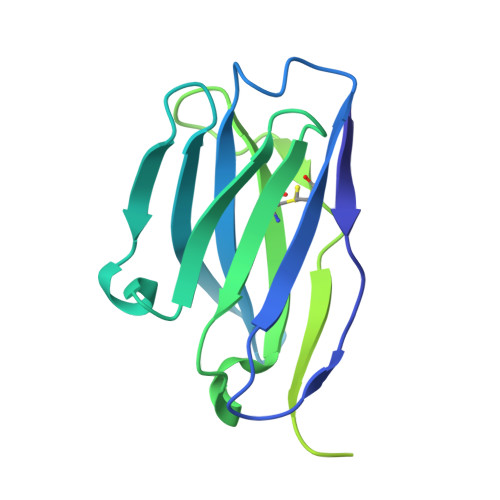
Nipah virus (NiV) and Hendra virus (HeV) have an extremely high case fatality, leading to hundreds of deaths in several countries around the globe. Belonging to the same genus Henipavirus (HNV), the two species have a high degree of sequence similarity, resulting in cross-neutralizing immunity under favorable conditions. Here, we obtained ten anti-NiV-F monoclonal antibodies using hybridoma technology, and verified that these antibodies had potent neutralizing activities against epidemic NiV strains from different regions using a pseudovirus assay, and the neutralizing concentration reached the nanogram per milliliter level. Moreover, two of the antibodies, NiF03-3C9 and NiF03-2F6, were found to have cross-neutralizing activity against HeV, which was even stronger than that against NiV. Epitope competition analysis revealed two classes of epitopes for these antibodies. Cryo-electron microscopy showed that NiF03-3C9 binds to lateral residues of the prefusion F protein trimer, highly conserved in both Nipah and Hendra. The protective potency of the antibodies was also validated using in vivo pseudovirus infection models of Nipah and Hendra viruses. The mAbs developed in this study and their conserved cross-neutralizing epitopes elucidated by structural analysis may contribute to the control of highly pathogenic HNV outbreaks.
- Division of HIV/AIDS and Sex-transmitted Virus Vaccines, Institute for Biological Product Control, National Institutes for Food and Drug Control (NIFDC), Beijing, 102629, China; State Key Laboratory of Drug Regulatory Science, Beijing, 102629, China.
Organizational Affiliation: