Novel oligomerization state of DinJ-YafQ complex from Vibrio cholerae-Structural and biochemical insights.
Mun, S.A., Yeon, G.B., Park, Y.H., Yoon, H., Ha, J.H., Jang, T.H., Hong, E., Bae, J., Son, J.(2025) Int J Biol Macromol 320: 145865-145865
- PubMed: 40639525
- DOI: https://doi.org/10.1016/j.ijbiomac.2025.145865
- Primary Citation of Related Structures:
9LEW - PubMed Abstract:
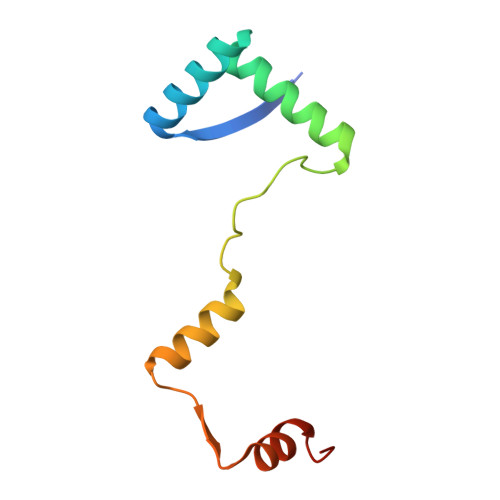
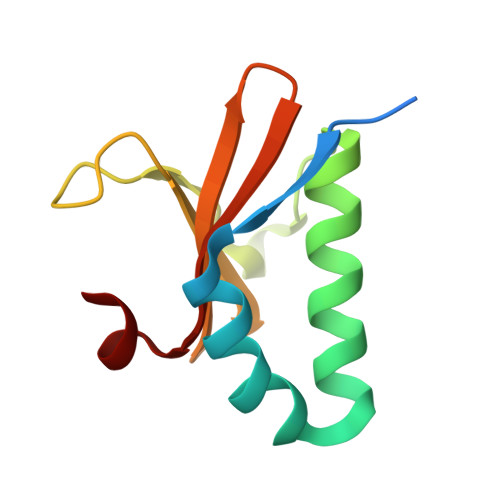
Toxin-antitoxin (TA) systems are widely distributed in archaeal and bacterial genomes and are crucial for maintaining the physiological functions required for cellular homeostasis. The DinJ-YafQ TA system belongs to the well-known RelBE family of Type II TA systems. Vibrio cholerae, the causative agent of cholera, infects humans through the consumption of contaminated, unpurified drinking water. Its genome contains a cassette of 19 TA system-related genes located on chromosome II, six of which are classified under the RelBE family. Herein, we determined the crystal structure of the V. cholerae DinJ (VcDinJ, antitoxin)-YafQ (VcYafQ, toxin) complex at 2.3 Å resolution. The unique orientation of the VcDinJ RHH 2 motifs relative to the YafQ molecules promotes the formation of a highly oligomerized hetero-hexadecamer (16-mer) in a concentration-dependent manner. According to biochemical experiments, the VcDinJ-YafQ complex directly binds to the promoter region of its operon, suggesting its role as a transcriptional repressor. An autoregulatory mechanism in which DNA binding induces conformational changes is suggested based on structural modeling through DNA superposition. This finding supports the concentration-dependent and DNA-associated oligomerization of the VcDinJ-YafQ complex.
- New Drug Development Center, Daegu-Gyeongbuk Medical Innovation Foundation (DGMIF), 360-4 Dongnae-dong, Dong-gu, Daegu 41061, Republic of Korea.
Organizational Affiliation: