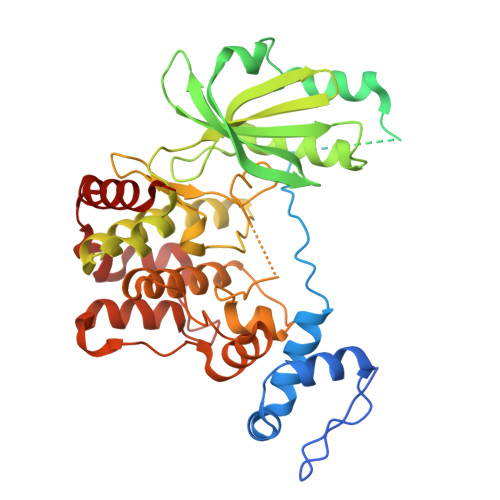
Crystal structures of PAK2 reveal new insights into its autoinhibitory mechanism.
Hu, H.F., Luo, Z., Zhang, Y., Fang, X., Zhu, Z., Wu, J.W., Wang, Z.X.(2025) Structure 33: 1663
- PubMed: 40752491
- DOI: https://doi.org/10.1016/j.str.2025.07.008
- Primary Citation of Related Structures:
9LBF, 9LBG, 9M41 - PubMed Abstract:
Type I p21-activated kinases (PAK1/2/3) exist in an auto-inhibited form and are stimulated by small G-protein binding and auto-phosphorylation. Previous structural and biochemical studies suggested that PAK1 is a dimer in crystals, and probably in a trans-inhibited conformation in solution. Here, we used multiple independent biochemical and biophysical methods to determine the oligomeric state and autoinhibitory mechanism of PAK2. Crystal structures of the full-length and N-terminal truncated PAK2 reveal the molecular basis underlying the PAK2 autoinhibition. Analytical ultracentrifugation studies show that these proteins have molecular weights that are consistent with monomeric species. The solution-phase structure of the full-length PAK2 by small angle X-ray scattering and computational modeling further shows a compact but elongated molecular shape. These results, taken together with the results of previous studies, demonstrate that in contrast with the most widely accepted model, all three type I PAKs are monomeric in solution and auto-inhibited in cis before activation.
- MOE Key Laboratory of Geriatric Diseases and Immunology, Institute of Molecular Enzymology, Soochow University, Suzhou, Jiangsu 215123, P.R. China; MOE Key Laboratory for Protein Sciences, School of Life Sciences, Tsinghua University, Beijing 100084, P.R. China.
Organizational Affiliation: