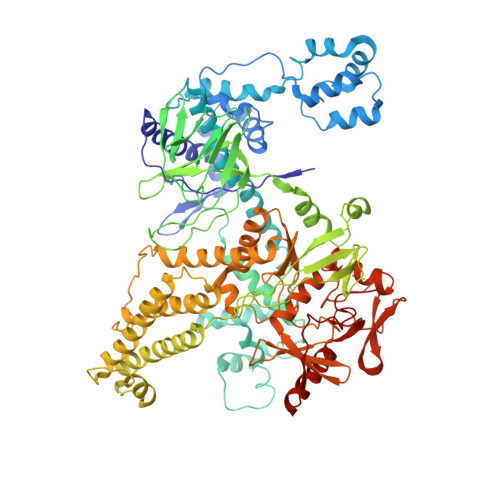
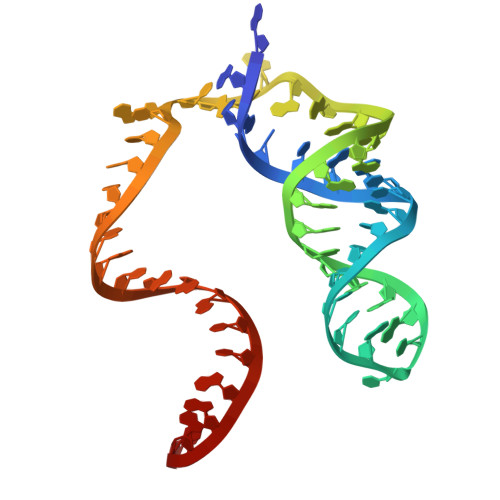
Cas12h is a crRNA-guided DNA nickase that can be utilized for precise gene editing.
Xiang, W., Lin, X., Yang, Y., Huang, L., Chen, Y., Chen, J., Liu, L.(2025) Cell Rep 44: 115718-115718
- PubMed: 40372912
- DOI: https://doi.org/10.1016/j.celrep.2025.115718
- Primary Citation of Related Structures:
9L12 - PubMed Abstract:
Type V-H CRISPR-Cas system, an important subtype of type V CRISPR-Cas systems, has remained enigmatic in terms of its structure and function despite being discovered several years ago. Here, we comprehensively characterize the type V-H CRISPR-Cas system and elucidate its role as a DNA nicking system. The unique CRISPR RNA (crRNA) employed by Cas12h effector protein enables specific targeting of double-stranded DNA (dsDNA), while its RuvC domain is responsible for cleaving the non-target strand (NTS) of dsDNA. We present the structure of Cas12h bound to crRNA and target DNA. Our structural analysis reveals that the RuvC domain possesses a narrow active pocket that facilitates recognition of NTS but potentially hinders access to the target strand. Furthermore, we demonstrate that Cas12h confers adaptive immunity against invading mobile genetic elements through transcriptional gene inhibition. We have engineered an adenine base editor by fusing Cas12h with an adenine deaminase, achieving effective A-to-G substitution.
- State Key Laboratory of Cellular Stress Biology, Xiang'an Hospital, School of Life Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiamen 361102, China.
Organizational Affiliation: