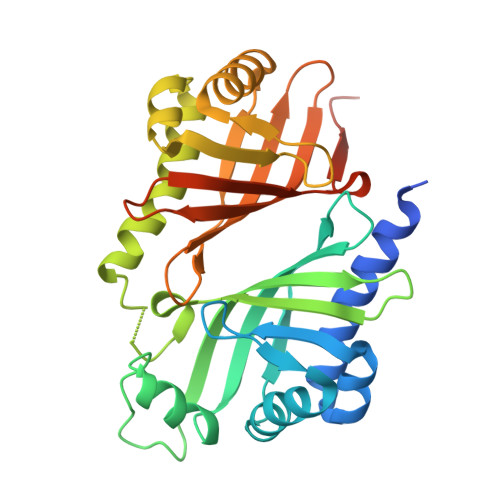
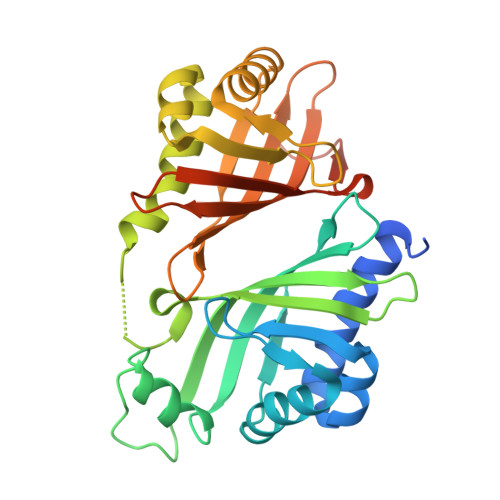
A paring system of polyether epoxide hydrolases enables a mouldable enzyme for consecutive ring cyclisation cascade
Yabuno, N., Minami, A., Ozaki, T., Owada, Y., Sawada, K., Arai, A., Tadokoro, T., Aizawa, T., Dosen, T., Nomai, T., Matsumaru, T., Junyang, L., Tao, Y., Kodama, A., Uchiyama, S., Hengphasatporn, K., Shigeta, Y., Saio, T., Maenaka, K., Yao, M., Kumeta, H., Oikawa, H., Ose, T.To be published.