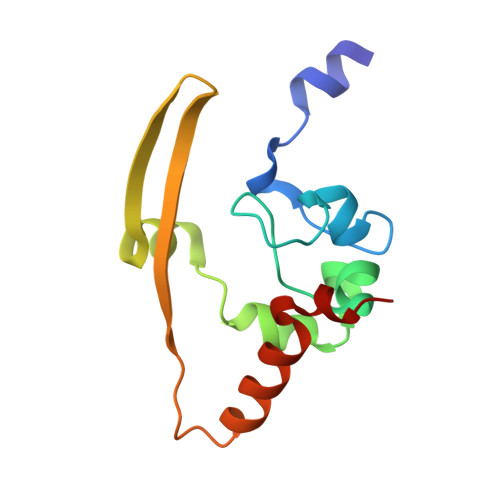
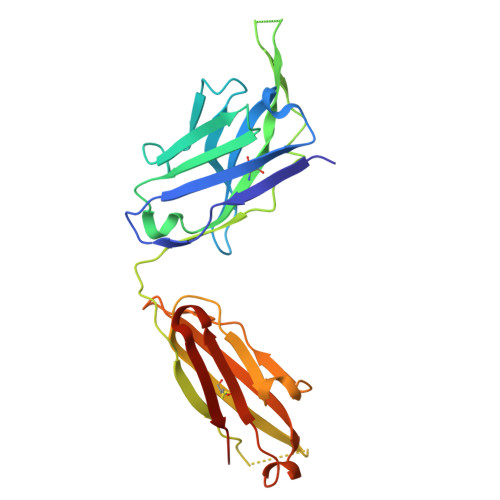
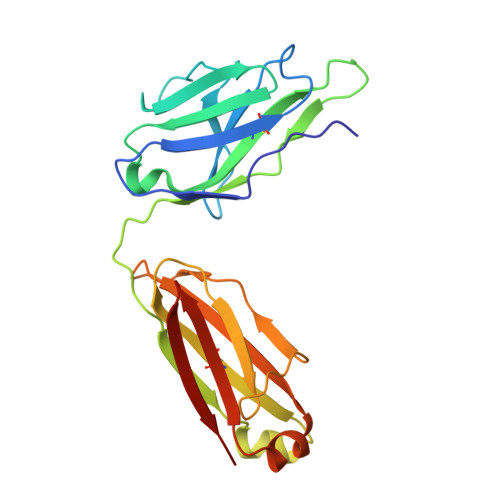
Structural basis of a human antibody targeting SARS-CoV-2 nucleocapsid protein dimerization domain and interfering with RNA-binding.
Xue, S., He, S., Huang, Z., Yang, M., Hu, G., Chen, X., Chen, Q., Zhou, W., Lin, S., Chen, S.(2025) Commun Biol 8: 1248-1248
- PubMed: 40830575
- DOI: https://doi.org/10.1038/s42003-025-08648-x
- Primary Citation of Related Structures:
9KUR - PubMed Abstract:
The transition of SARS-CoV-2 into a recurrent, seasonal pathogen has underscored the need for the induction of durable immune protection. The nucleocapsid (N) protein is regarded as a promising complementary target for therapeutic and vaccine strategies, owing to its structural robustness, clinical relevance, and ability to elicit critical immune response. Within the N protein, the C-terminal domain (N-CTD) plays a pivotal role in assembly of viral RNA (vRNA)-N protein complexes, and in facilitating liquid-liquid phase separation (LLPS) through specific interactions with RNA on its dimerization surface. Despite its functional importance, the molecular mechanisms by which the RNA-binding surface of this domain can be selectively targeted by inhibitors remain poorly defined. Herein, we report a 2.06 Å crystal structure of the SARS-CoV-2 N-CTD in complex with nCoV400Fab, a human monoclonal antibody derived from single B-cell screening. The structure reveals that nCoV400Fab engages multiple basic residues on the RNA-binding surface, forming a steric blockade that hinders vRNA binding. Functional assays demonstrate that nCoV400Fab disrupts both viral ribonucleoprotein (vRNP) assembly and RNA-induced condensate formation. Together, these findings define a structural mechanism by which a human antibody disrupts the RNA-binding surface of N-CTD, laying the foundation for the development of macromolecular inhibitors.
- Molecular Imaging Center, The Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai, Guangdong, China.
Organizational Affiliation: