Human Hsp90-Cdc37-PINK1 complex
Tian, X.Y., Su, J.Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Heat shock protein HSP 90-alpha | 732 | Homo sapiens | Mutation(s): 0 EC: 3.6.4.10 | 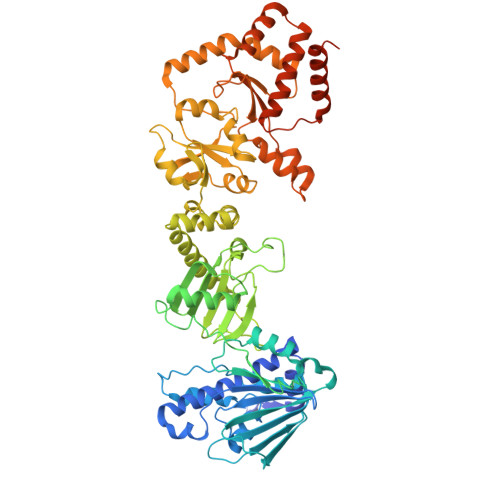 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P07900 (Homo sapiens) Explore P07900 Go to UniProtKB: P07900 | |||||
PHAROS: P07900 GTEx: ENSG00000080824 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P07900 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Serine/threonine-protein kinase PINK1, mitochondrial | 494 | Homo sapiens | Mutation(s): 0 Gene Names: PINK1 EC: 2.7.11.1 | 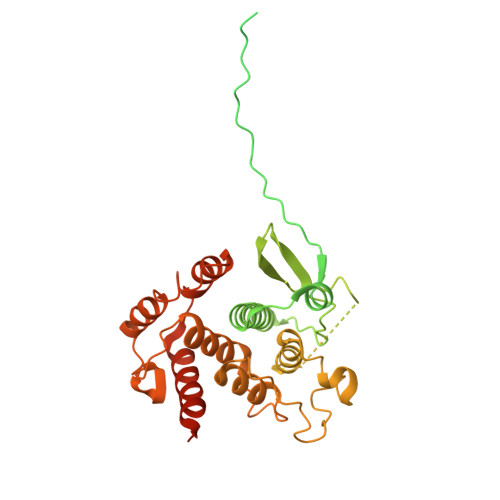 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9BXM7 (Homo sapiens) Explore Q9BXM7 Go to UniProtKB: Q9BXM7 | |||||
PHAROS: Q9BXM7 GTEx: ENSG00000158828 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9BXM7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hsp90 co-chaperone Cdc37 | D [auth E] | 378 | Homo sapiens | Mutation(s): 0 | 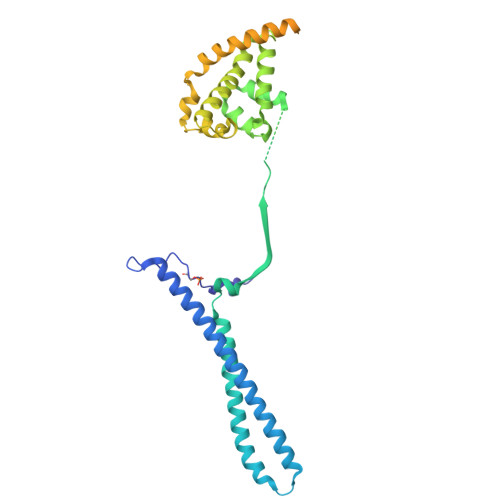 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q16543 (Homo sapiens) Explore Q16543 Go to UniProtKB: Q16543 | |||||
PHAROS: Q16543 GTEx: ENSG00000105401 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q16543 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP (Subject of Investigation/LOI) Query on ADP | E [auth A], F [auth B] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| SEP Query on SEP | D [auth E] | L-PEPTIDE LINKING | C3 H8 N O6 P |  | SER |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32071192 |