Structural basis of nectin-4 recognition by the antibody-drug conjugate 9MW2821.
Fang, P., You, M., Wen, H., Cao, Y., Zhou, W., Zhu, X., Shi, L., Sun, X., Li, K., Li, W., Wang, J., Wu, H., Tan, X.(2025) J Biological Chem 301: 110816-110816
- PubMed: 41101504
- DOI: https://doi.org/10.1016/j.jbc.2025.110816
- Primary Citation of Related Structures:
9KKJ - PubMed Abstract:
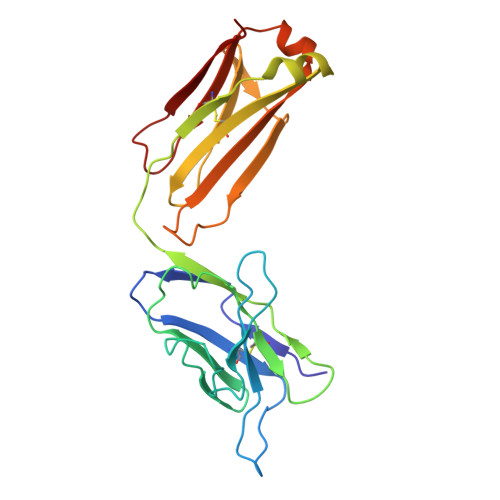
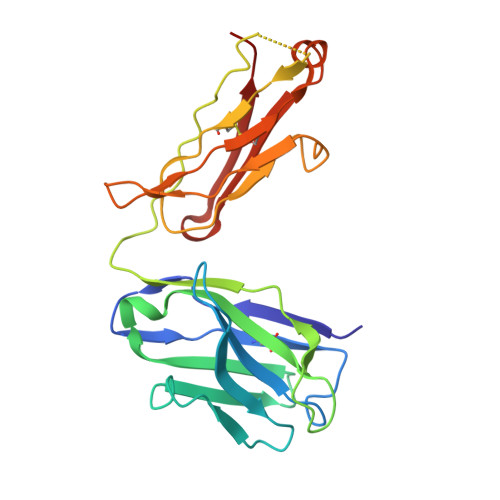
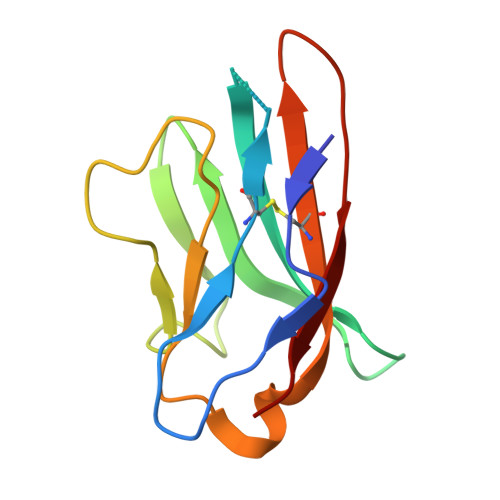
Nectin-4, a membrane protein highly expressed in multiple solid tumors, has become an attractive target for antibody-drug conjugate development. We designed and developed 9MW2821, an anti-nectin-4 antibody-drug conjugate with an enzymatically cleavable valine-citrulline linker and monomethyl auristatin E as the payload. Although 9MW2821 has shown good efficacy and safety in multiple solid tumors in clinical trials, the interaction between 9MW2821 and nectin-4 at the molecular level remains unclear. In this study, we solved the structure of the antigen-binding fragment of 9MW2821 in complex with nectin-4 at a resolution of 3.26 Å using single-particle cryo-EM. The structure shows that 9MW2821 binds the front β-sheet of nectin-4 D1 through three complementarity-determining region loops from the heavy chain and two complementarity-determining region loops from the light chain. The binding involves extensive hydrogen bonds and hydrophobic interactions. The buried surface area is more than 1600 Å 2 . Mutagenesis studies revealed that four residues (Q77, E78, H83, and E95) of nectin-4 contributed significantly to the binding of 9MW2821 monoclonal antibody (mAb). The structure also shows that 9MW2821 mAb blocks nectin-4 homodimer formation by competing with the nectin-4 partner D1 domain for part of the nectin-4 surface area. Furthermore, 9MW2821 mAb prevents nectin-4 from interacting with nectin-1 and T-cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domain, enhancing the activation of NK cells. Based on the structure of the nectin-4 D1-9MW2821 antigen-binding fragment complex resolved in this study, we have elucidated the molecular mechanism of 9MW2821 in cancer therapy. The findings provide a basis for optimizing future mAbs targeting nectin-4.
- Department of Quality, Jiangsu Mabwell Health Pharmaceutical R&D Co., Ltd., Taizhou, Jiangsu Province, China.
Organizational Affiliation: