Molecular mechanism underlying non-discriminatory recognition of relaxin-3 by RXFP3 and RXFP4.
Chen, Y., Zhou, Q., Yan, S., Yan, J., Yang, D., Chen, J., Wang, M.W.(2025) Commun Biol 8: 794-794
- PubMed: 40410443
- DOI: https://doi.org/10.1038/s42003-025-08220-7
- Primary Citation of Related Structures:
9KFI, 9KFJ, 9KFK - PubMed Abstract:
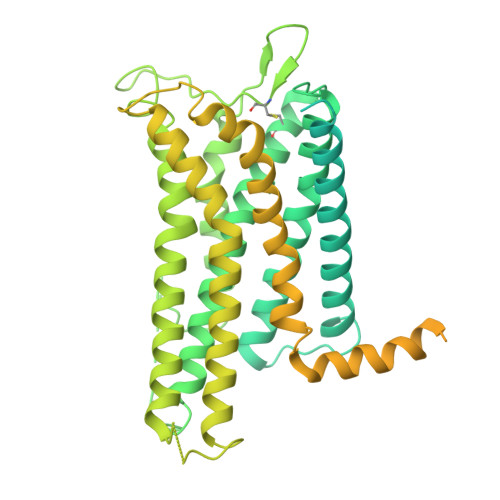
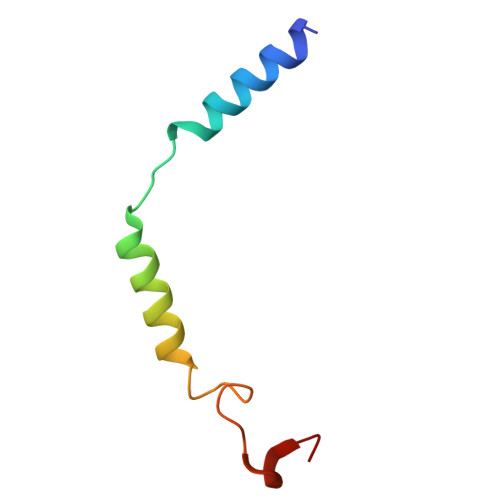
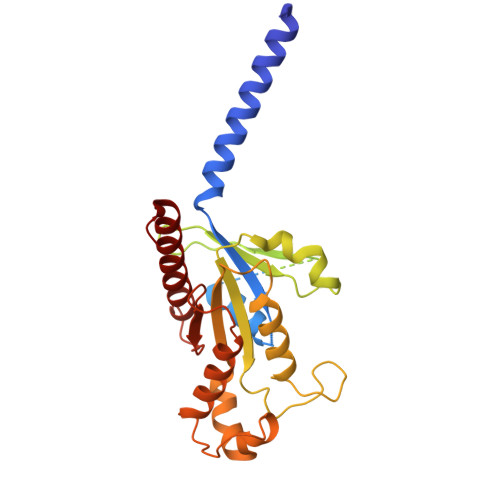
The human relaxin family peptide receptors RXFP3 and RXFP4 play important physiological roles through interactions with endogenous hormones, relaxin-3 and insulin-like peptide 5 (INSL5). They are implicated in certain neurological and metabolic disorders. While INSL5 only activates RXFP4, relaxin-3 is recognized by both receptors. Here, we report the cryo-electron microscopy structures of RXFP3-G i complexes bound by relaxin-3 or a small-molecule dual agonist (compound 4), and relaxin-3 in complex with RXFP4-G i , with global resolutions of 2.91 Å, 2.95 Å, and 3.10 Å, respectively. It is found that relaxin-3 adopts a conserved binding conformation within the transmembrane domain (TMD) bundle of RXFP3 and RXFP4, where the C-terminal tip residues of its B chain, R26 and W27, make extensive contacts with conserved receptor residues, thereby activating RXFP3 and RXFP4. Compound 4 mimics these key interactions by binding to both receptors. In contrast, the C-terminal residues composition and TMD-binding angle of INSL5 in RXFP4 differ significantly from that of relaxin-3, ensuring its selectivity for RXFP4. These findings deepen our understanding about the structural basis of ligand recognition and selectivity in this G protein-coupled receptor subfamily.
- Research Center for Medicinal Structural Biology, National Research Center for Translational Medicine at Shanghai, State Key Laboratory of Medical Genomics, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Organizational Affiliation: