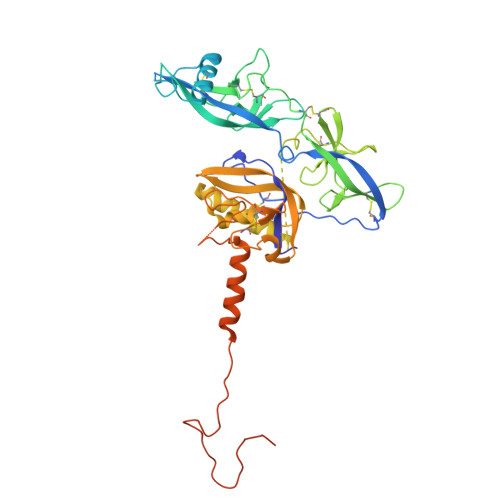
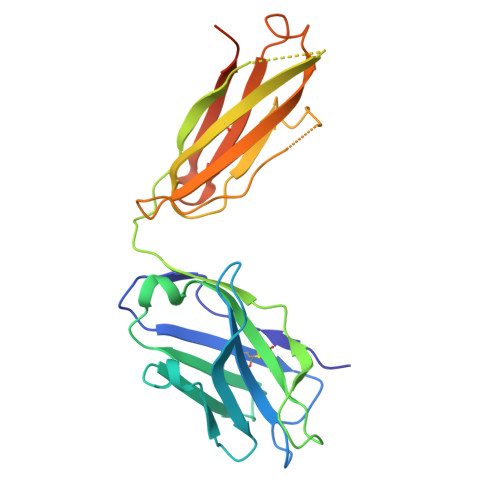
Molecular basis of two broad-spectrum antibodies neutralizing rabies virus and other phylogroup-I lyssaviruses by blocking structural transition between the pleckstrin-homology and fusion domains in the glycoprotein.
Yang, F., Zhai, L., Yin, K., Lin, S., Yang, J., Ye, F., Chen, Z., Shu, S., Yu, Y., Guo, L., He, B., Wang, W., Ye, H., Cao, Y., Gao, J., Lu, G.(2025) Int J Biol Macromol 308: 142570-142570
- PubMed: 40154685
- DOI: https://doi.org/10.1016/j.ijbiomac.2025.142570
- Primary Citation of Related Structures:
9KEF, 9KEP, 9KFB - PubMed Abstract:
Rabies virus (RABV), the prototype species of the Lyssavirus genus, causes the lethal disease of rabies. Rabies can be efficiently prevented with post-exposure prophylaxis. The RABV glycoprotein (RABV-G) is an essential factor mediating virus entry and the major target of neutralizing antibodies. Here, we report the crystal structures of two neutralizing monoclonal antibodies (NM57 and SOJB) in complex with RABV-G. The two antibodies recognize highly overlapped epitopes involving both the pleckstrin-homology domain (PHD) and the junction between PHD and the fusion domain (FD). Our pseudovirus neutralization assay further shows that both antibodies could neutralize a majority of the lyssaviruses in phylogroup-I. Via sequence comparison and structural characterization, we identify two residues located at positions 226 and 231 in PHD, as key determinants for antibody recognition, which is further corroborated by mutagenesis analyses. Finally, detailed structural analyses reveal that NM57 and SOJB would lock the PHD/FD local structure in the pre-fusion-like state, thereby inhibiting viral infection by blocking structural transitions of RABV-G essential for membrane fusion. Taken together, these results provide a mechanistic glimpse into the molecular basis for broad neutralization of phylogroup-I lyssaviruses by NM57 and SOJB, which should be able to facilitate the development of monoclonal antibodies and vaccines.
- Department of Emergency Medicine, State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, Sichuan 610041, China. Electronic address: yfanli16@qq.com.
Organizational Affiliation: