Structural basis of a sodium pump quaternary complex leading phospholipid scrambling in the living cell
Dou, Y., Abe, K., Suzuki, J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sodium/potassium-transporting ATPase subunit alpha-1 | 985 | Homo sapiens | Mutation(s): 0 Gene Names: ATP1A1 EC: 7.2.2.13 | 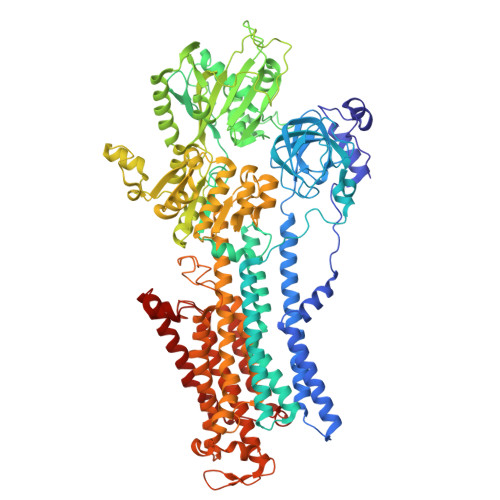 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P05023 (Homo sapiens) Explore P05023 Go to UniProtKB: P05023 | |||||
PHAROS: P05023 GTEx: ENSG00000163399 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P05023 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sodium/potassium-transporting ATPase subunit beta-3 | 279 | Homo sapiens | Mutation(s): 0 Gene Names: ATP1B3 | 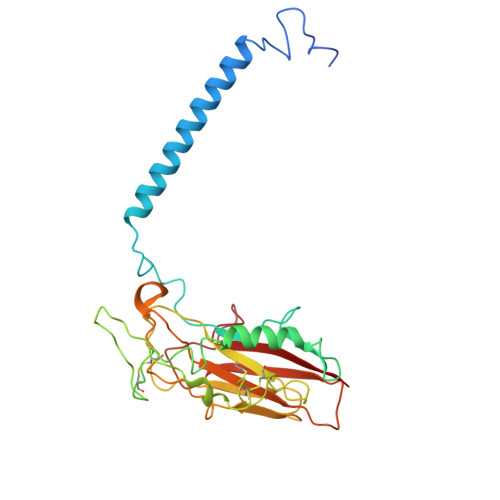 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P54709 (Homo sapiens) Explore P54709 Go to UniProtKB: P54709 | |||||
PHAROS: P54709 GTEx: ENSG00000069849 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P54709 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| FXYD domain-containing ion transport regulator 5 | 178 | Homo sapiens | Mutation(s): 0 Gene Names: FXYD5, DYSAD, IWU1, HSPC113, UNQ2561/PRO6241 | 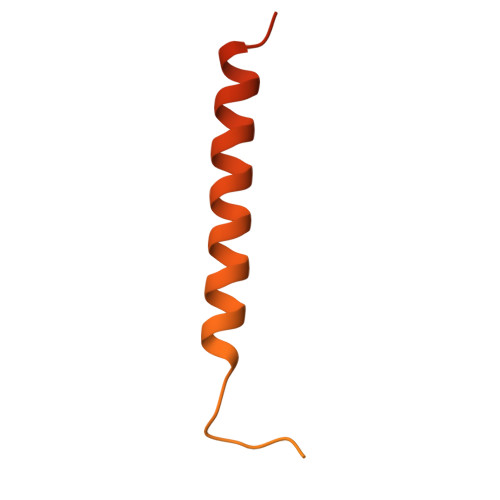 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96DB9 (Homo sapiens) Explore Q96DB9 Go to UniProtKB: Q96DB9 | |||||
PHAROS: Q96DB9 GTEx: ENSG00000089327 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96DB9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PC1 Query on PC1 | J [auth A], N [auth B] | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE C44 H88 N O8 P NRJAVPSFFCBXDT-HUESYALOSA-N |  | ||
| PCW Query on PCW | L [auth A], M [auth B] | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE C44 H85 N O8 P SNKAWJBJQDLSFF-NVKMUCNASA-O |  | ||
| ACP (Subject of Investigation/LOI) Query on ACP | I [auth A] | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER C11 H18 N5 O12 P3 UFZTZBNSLXELAL-IOSLPCCCSA-N |  | ||
| CLR Query on CLR | H [auth A], K [auth A], O [auth B] | CHOLESTEROL C27 H46 O HVYWMOMLDIMFJA-DPAQBDIFSA-N |  | ||
| MG Query on MG | G [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| NA (Subject of Investigation/LOI) Query on NA | D [auth A], E [auth A], F [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Society for the Promotion of Science (JSPS) | Japan | 21H02426 |