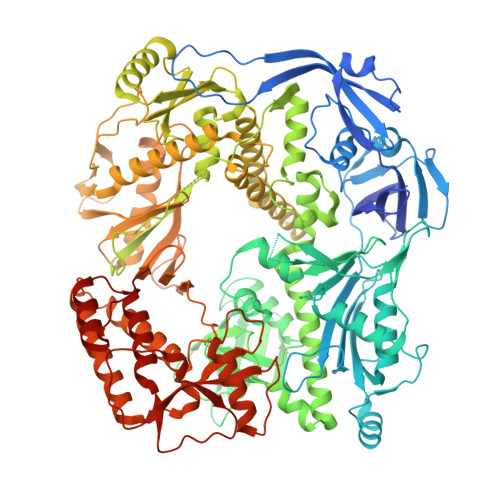
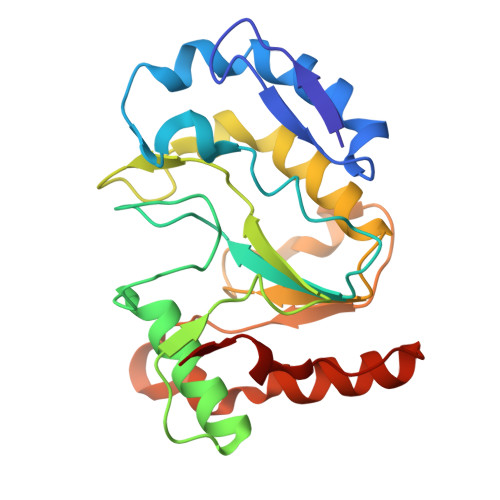
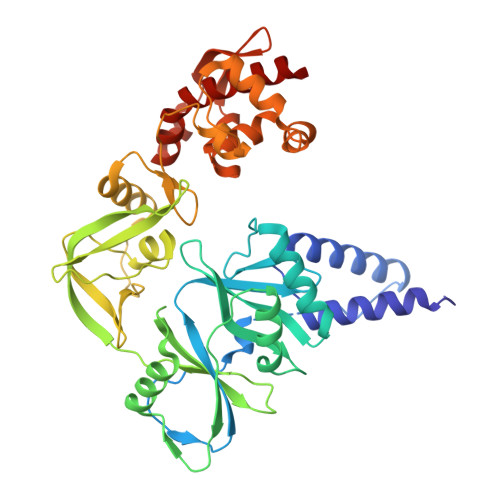
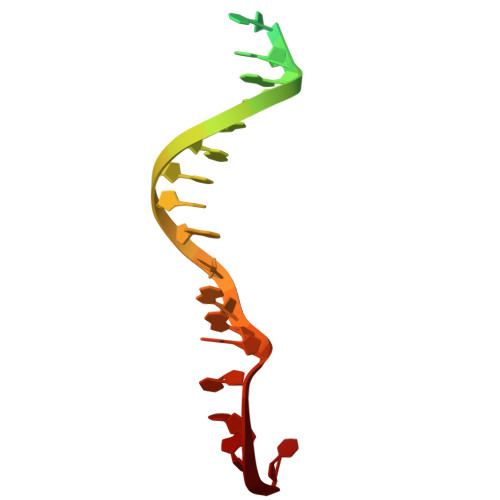
Structural basis of DNA replication fidelity of the Mpox virus.
Xie, Y., Kuai, L., Peng, Q., Wang, Q., Wang, H., Li, X., Qi, J., Ding, Q., Shi, Y., Gao, G.F.(2025) Proc Natl Acad Sci U S A 122: e2411686122-e2411686122
- PubMed: 40035768
- DOI: https://doi.org/10.1073/pnas.2411686122
- Primary Citation of Related Structures:
9K9R, 9K9S, 9K9T, 9K9U, 9K9V - PubMed Abstract:
The Mpox virus (MPXV) is an orthopoxvirus that caused a global outbreak in 2022. The poxvirus DNA polymerase complex is responsible for the replication and integrity of the viral genome; however, the molecular mechanisms underlying DNA replication fidelity are still unclear. In this study, we determined the cryoelectron microscopy (cryo-EM) structures of the MPXV F8-A22-E4 polymerase holoenzyme in its editing state, in complex with mismatched primer-template DNA and DNA containing uracil deoxynucleotide. We showed that the MPXV polymerase has a similar replication-to-edit transition mechanism to proofread the mismatched nucleotides like the B-family DNA polymerases of other species. The unique processivity cofactor A22-E4 undergoes conformational changes in different working states and might affect the proofreading process. Moreover, we elucidated the base excision repair (BER) function of E4 as a uracil-DNA glycosylase and the coupling mechanism of genome replication and BER, characteristic of poxviruses. Our findings greatly enhance our molecular understanding of DNA replication fidelity of orthopoxviruses and will stimulate the development of broad-spectrum antiviral drugs.
- Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: