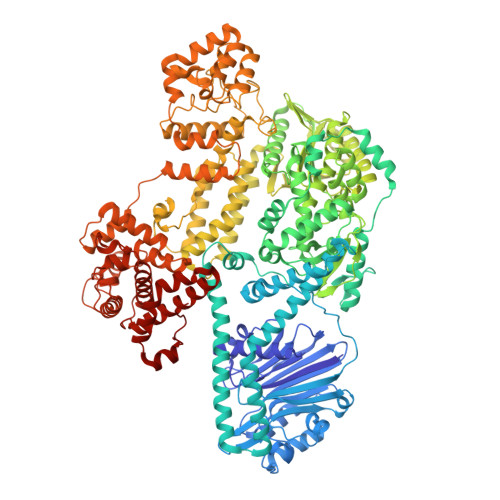
Mechanism of DNA targeting by human LINE-1.
Jin, W., Yu, C., Zhang, Y., Cao, C., Xia, T., Song, G., Cai, Z., Xue, Y., Zhu, B., Xu, R.M.(2025) Science 390: eadu3433-eadu3433
- PubMed: 41066570
- DOI: https://doi.org/10.1126/science.adu3433
- Primary Citation of Related Structures:
9K6G, 9K6H, 9K6I - PubMed Abstract:
Long interspersed nuclear element-1 (LINE-1 or L1), the only autonomously active retrotransposon in humans today, constitutes a large proportion of the genome and continues to evolve the genome and impact fundamental biological processes. L1 retrotransposition critically depends on its endonuclease and reverse transcriptase subunit open reading frame 2 protein (ORF2p), which targets genomic loci and nicks DNA using an evolutionarily distinct yet not fully understood mechanism. Our structural and biochemical analyses revealed that ORF2p is a structure-dependent endonuclease. It binds a double-stranded DNA region upstream of the nicking site and recognizes a downstream forked or flap structure for efficient DNA nicking. This discovery suggests that L1 mobilization piggybacks on chromosomal processes with noncanonical DNA structure intermediates.
- State Key Laboratory of Epigenetic Regulation and Intervention, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: