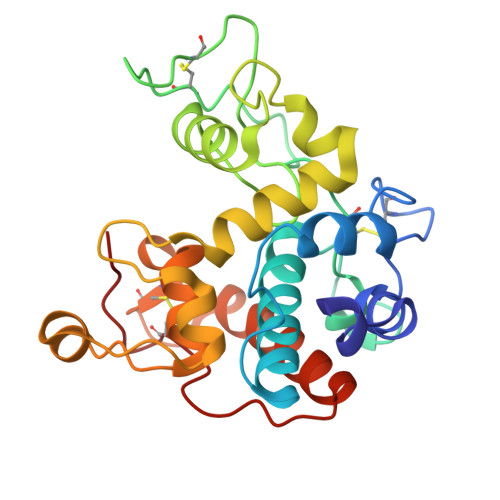
Structural and functional characterization of chitinase from carnivorous plant Drosera adelae.
Yoneda, K., Naruse, Y., Suizu, Y., Araki, T., Hoshi, Y., Sakuraba, H., Hayashi, J., Ohshima, T.(2025) FEBS Open Bio 15: 1930-1944
- PubMed: 40874485
- DOI: https://doi.org/10.1002/2211-5463.70110
- Primary Citation of Related Structures:
9JTP, 9JTR - PubMed Abstract:
A class I chitinase from the carnivorous sundew plant Drosera adelae was successfully expressed in the methylotrophic yeast Pichia pastoris and efficiently purified using a chitin affinity column. Enzymatic activity assays revealed that the enzyme showed a specific activity of 235.3 ± 10.2 U·mg -1 . Crystallization of wild-type and E167Q catalytic mutant chitinases yielded needle-like microcrystals. X-ray diffraction experiments were performed, and high-resolution datasets were obtained at 1.73 Å and 1.57 Å, respectively. Structural analysis of diffraction data revealed that only the catalytic domain could be resolved in both crystal forms. Using AutoDock Vina, we performed docking simulations of two (GlcNAc) 4 molecules at eight subsites (+4 to -4) of the catalytic domain of D. adelae chitinase to investigate their binding energies and conformations. Further, the structure of the chitin-binding domain (hevein domain), which could not be resolved by X-ray crystallography, was predicted using alphafold2. Based on this model, the binding conformation and binding energy of (GlcNAc) 3 were analyzed using similar methods. In D. adelae chitinase, a characteristic tyrosine cluster consisting of Tyr174, Tyr199, and Tyr201 formed a unique structural feature that enabled recognition of the (GlcNAc) 4 substrate. The hevein domain structures further indicated that the tyrosine cluster (Tyr41, Tyr43, Tyr50) in D. adelae chitinase may be involved in hydrogen bonding and CH/π interactions with (GlcNAc) 3 .
- Department of Food and Life Sciences, School of Agriculture, Tokai University, Kumamoto, Japan.
Organizational Affiliation: