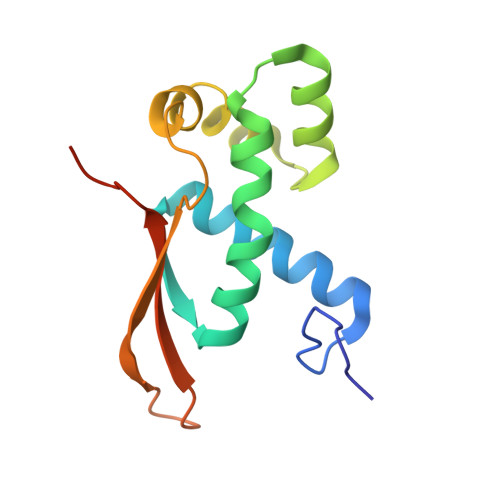
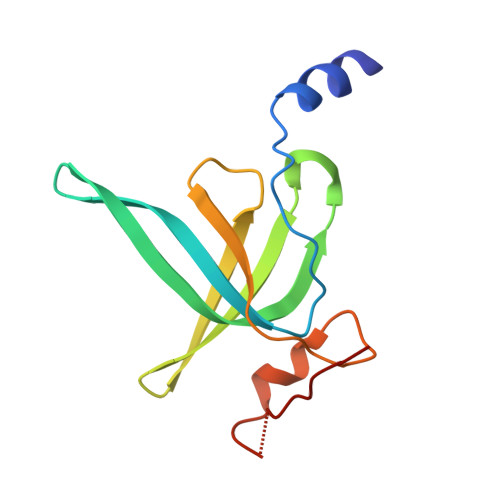
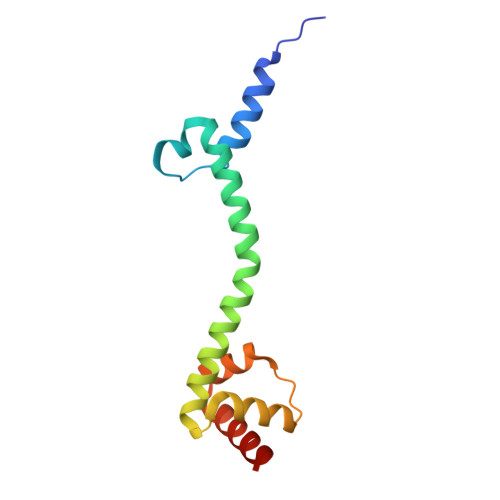
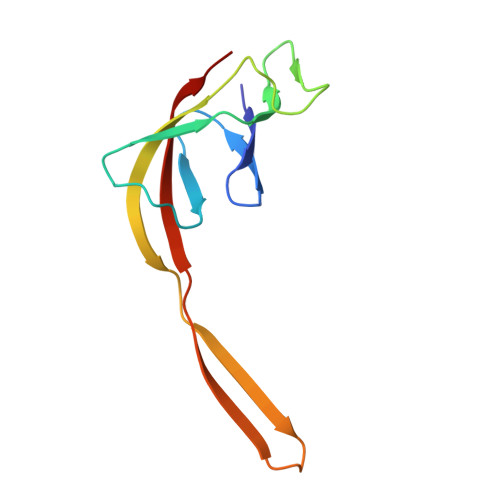
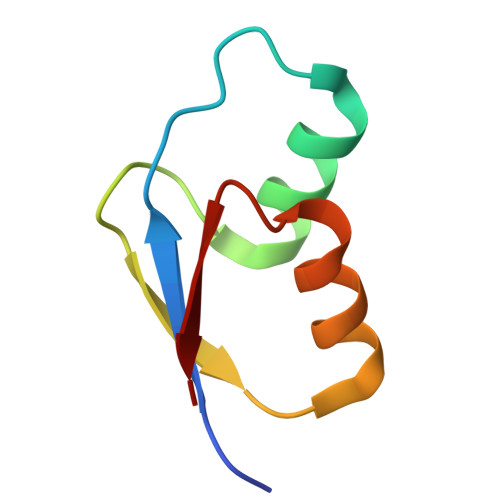
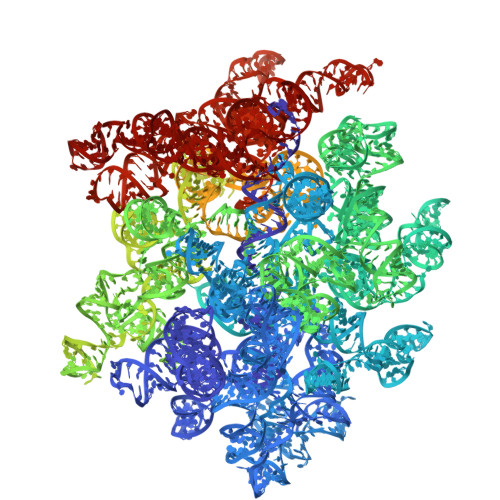
Mechanistic insights into 50 S precursor recognition and targeting by erythromycin resistance methyltransferase.
Sengupta, S., Mukherjee, R., Pilsl, M., Bagale, S., Adhikary, A.D., Borkar, A.N., Pradeepkumar, P.I., Engel, C., Chowdhury, A., Kaushal, P.S., Anand, R.(2025) Sci Adv 11: eaea1545-eaea1545
- PubMed: 41296849
- DOI: https://doi.org/10.1126/sciadv.aea1545
- Primary Citation of Related Structures:
9JMK, 9JNS, 9JSR - PubMed Abstract:
Erythromycin resistance methyltransferases (Erms) confer resistance to macrolide, lincosamide, and streptogramin B antibiotics by methylating an internal base (A2058, E. coli numbering) in an elusive precursor ribosomal state. Here, we capture the 50 S ribosomal precursor-Erm complex by cryo-EM and show that a transient pocket formed in the early steps of ribosome biogenesis, situated 35 angstrom from the methylation site, serves as an anchor for the auxiliary C-terminal domain of Erm, thereby playing a crucial role in achieving specificity in this short-lived substrate with evolving structural features. Cryo-EM reveals that the catalytic Rossman fold of Erm undergoes a swaying motion to facilitate substrate scouting. Corroboratory smFRET studies show that for effective catalysis, Erm transitions between multiple conformations, an effective strategy adopted to orient the dynamic helix where methylation occurs. Unraveling this unique mechanism of targeting adopted by Erm paves the way for selective design of allosteric inhibitors directed toward reversing MLS B resistance.
- Department of Chemistry, Indian Institute of Technology Bombay, Powai, Mumbai 400 076, Maharashtra, India.
Organizational Affiliation: