Structural insights into DdCBE in action enable high-precision mitochondrial DNA editing.
Xiang, J., Xu, W., Wu, J., Luo, Y., Liu, C., Hou, Y., Chen, J., Yang, B.(2025) Mol Cell 85: 3357
- PubMed: 40934924
- DOI: https://doi.org/10.1016/j.molcel.2025.08.016
- Primary Citation of Related Structures:
9JO8, 9KY4 - PubMed Abstract:
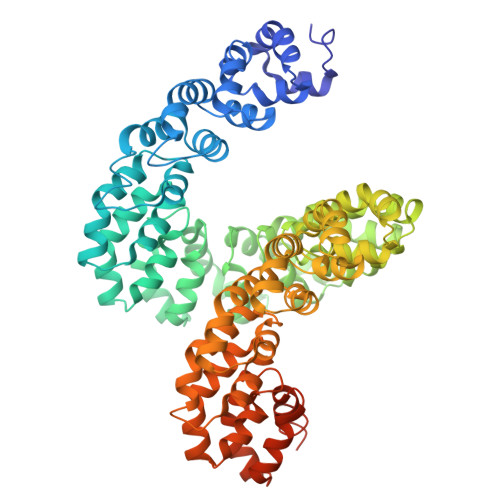
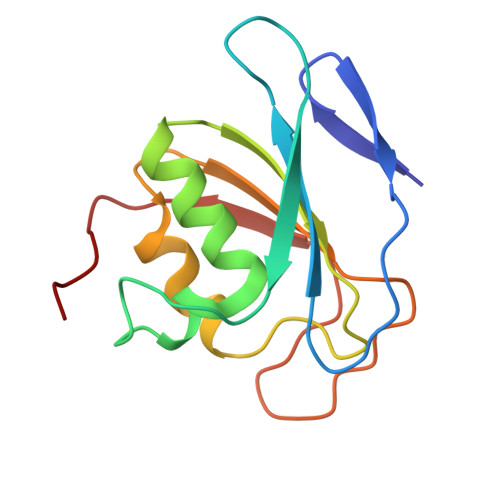
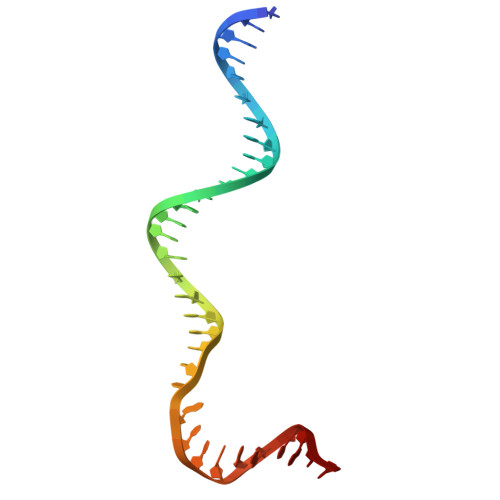
DddA-derived cytosine base editor (DdCBE) couples transcription activator-like effector (TALE) arrays and the double-stranded DNA (dsDNA)-specific cytidine deaminase DddA to target mitochondrial DNA (mtDNA) for editing. However, structures of DdCBE in action are unavailable, impeding its mechanistic-based optimization for high-precision-demanding therapeutic applications. Here, we determined the cryo-electron microscopy (cryo-EM) structures of DdCBE targeting two native mitochondrial gene loci and combined editing data from systematically designed spacers to develop WinPred, a model that can predict DdCBE's editing outcome and guide its design to achieve high-precision editing. Furthermore, structure-guided engineering of DddA narrowed the editing window of DdCBE to 2-3 nt while minimizing its off-target (OT) editing to near-background levels, thereby generating accurate DdCBE (aDdCBE). Using aDdCBE, we precisely introduced a Leber hereditary optic neuropathy (LHON)-disease-related mutation into mtDNA and faithfully recapitulated the pathogenic conditions without interference from unintended bystander or OT mutations. Our work provides a mechanistic understanding of DdCBE and establishes WinPred and aDdCBE as useful tools for faithfully modeling or correcting disease-related mtDNA mutations.
- Gene Editing Center, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Organizational Affiliation: