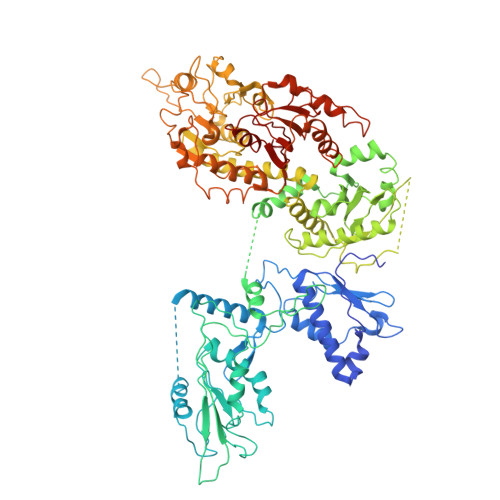
Structural and functional characterization of human SLFN14.
Luo, M., Jia, X., Wang, Z.W., Yang, J.Y., Wang, W., Chen, J., Ou, J.Y., Feng, J.X., Yu, B., Wang, S., Huang, L., Morgan, N.V., Deng, K., Chen, T., Zhang, Q., Gao, S.(2025) Nucleic Acids Res 53
- PubMed: 40464691
- DOI: https://doi.org/10.1093/nar/gkaf484
- Primary Citation of Related Structures:
9JN9 - PubMed Abstract:
The Schlafen (SLFN) family of proteins are a group of DNA/RNA processing enzymes with emerging importance in human health and disease, where their functions are implicated in a variety of immunological and anti-tumor processes. Here, we present the cryo-electron microscopy structure of full-length human SLFN14, a member with antiviral activity and linked to an inherited bleeding disorder. SLFN14 is composed of an RNase domain, a SWADL domain, and a two-lobe helicase domain. SLFN14 exhibited strong RNase activity over different substrates, and the positively charged patches at the valley of the RNase domain, which contains the thrombocytopenia-related missense mutation sites, are crucial for binding oligonucleotides. SLFN14 lacks helicase activity, which can be attributed to the inability to bind ATP and the absence of positive charges at the canonical DNA-binding site of its RecA-like folds. SLFN14 is structurally similar to SLFN11, but differs from SLFN5 in the orientation of the helicase domain. Live-cell fluorescence resonance energy transfer (FRET) assays and AlphaFold2 analysis hinted that SLFN14 may adopt multiple conformations in cells. These results provide detailed structural and biochemical features of SLFN14, and greatly expand our knowledge of the functional diversity of the SLFN family.
- State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou 510060, China.
Organizational Affiliation: