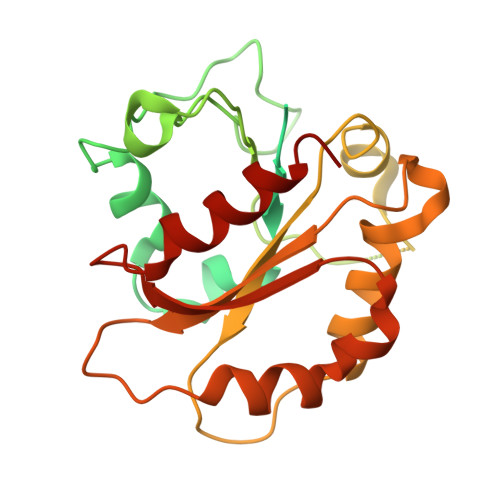
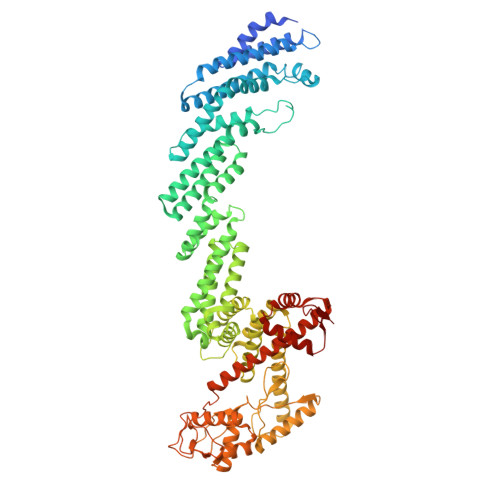
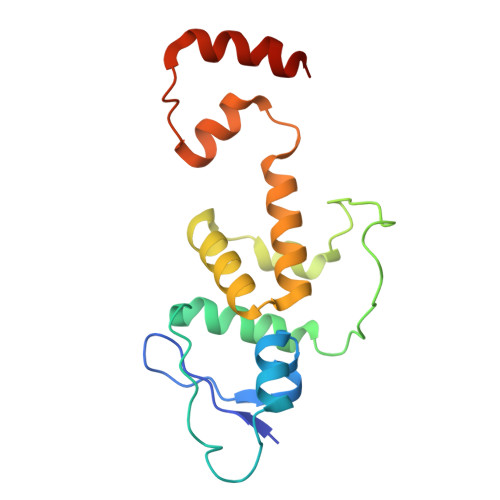
Structure of the CUL1-RBX1-SKP1-FBXO4 SCF ubiquitin ligase complex.
Zhu, W., Chen, X., Zhang, J., Xu, C.(2024) Biochem Biophys Res Commun 735: 150811-150811
- PubMed: 39406020
- DOI: https://doi.org/10.1016/j.bbrc.2024.150811
- Primary Citation of Related Structures:
9JKB - PubMed Abstract:
Cullin-RING E3 ubiquitin ligases (CRLs) constitute the largest family of ubiquitin ligase and play important roles in regulation of proteostasis. Here we presented the cryo-EM structure of CRL1 FBXO4 , a member of Cullin-1 E3 ligase. CRL1 FBXO4 adopts a homodimer architecture. Structural analysis revealed that in the CRL1 FBXO4 protomer, the substrate recognition subunit FBXO4 interacts both the adaptor protein SKP1, and the scaffold protein CUL1 via hydrophobic and electrostatic interactions. Two FBXO4 forms a domain-swapped dimer in the CRL1 FBXO4 structure, which constitutes the basis for the dimerization of CRL1 FBXO4 . Inspired by the cryo-EM density, we modeled the architecture of whole CRL1 FBXO4 as a symmetrical dimer, which provides insights into CRL1 FBXO4 -medaited turnover of oncogene proteins.
- MOE Key Laboratory for Membraneless Organelles & Cellular Dynamics, Center for Advanced Interdisciplinary Science and Biomedicine of IHM, Hefei National Laboratory for Physical Sciences at the Microscale, Division of Life Sciences and Medicine, University of Science and Technology of China, 230027, Hefei, PR China.
Organizational Affiliation: