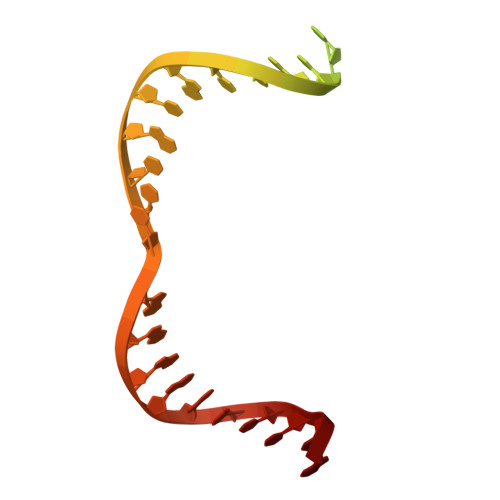Functional RNA splitting drove the evolutionary emergence of type V CRISPR-Cas systems from transposons.
Jin, S., Zhu, Z., Li, Y., Zhang, S., Liu, Y., Li, D., Li, Y., Luo, Y., Cheng, Z., Zhao, K.T., Gao, Q., Yang, G., Li, H., Liang, R., Zhang, R., Qiu, J.L., Zhang, Y.E., Liu, J.G., Gao, C.(2025) Cell 188: 6283-6300.e22
- PubMed: 41027434
- DOI: https://doi.org/10.1016/j.cell.2025.09.004
- Primary Citation of Related Structures:
9JFO, 9JFP, 9JFQ - PubMed Abstract:
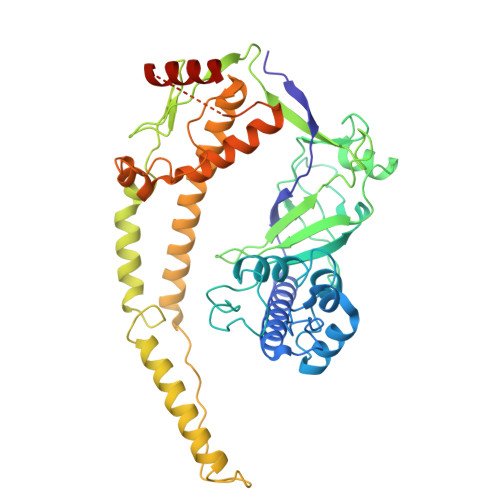
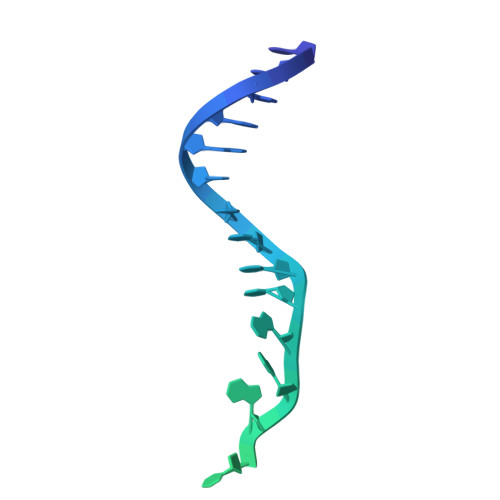
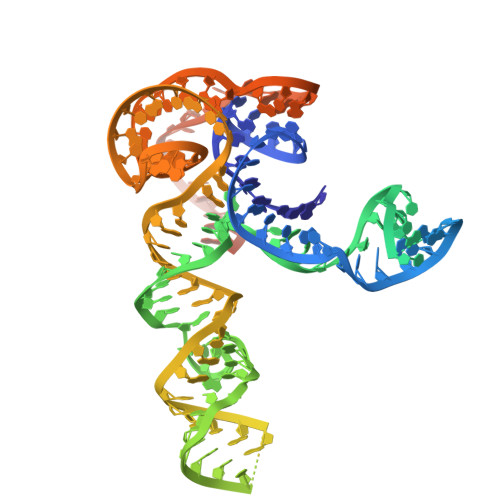
Transposon-encoded TnpB nucleases gave rise to type V CRISPR-Cas12 effectors through multiple independent domestication events. These systems use different RNA molecules as guides for DNA targeting: transposon-derived right-end RNAs (reRNAs or omega RNAs) for TnpB and CRISPR RNAs for type V CRISPR-Cas systems. However, the molecular mechanisms bridging transposon activity and CRISPR immunity remain unclear. We identify TranCs (transposon-CRISPR intermediates) derived from distinct IS605- or IS607-TnpB lineages. TranCs utilize both CRISPR RNAs and reRNAs to direct DNA cleavage. The cryoelectron microscopy (cryo-EM) structure of LaTranC from Lawsonibacter sp. closely resembles that of the ISDra2 TnpB complex; however, unlike a single-molecule reRNA, the LaTranC guide RNA is functionally split into a tracrRNA and crRNA. An engineered RNA split of ISDra2 TnpB enabled activity with a CRISPR array. These findings indicate that functional RNA splitting was the primary molecular event driving the emergence of diverse type V CRISPR-Cas systems from transposons.
- New Cornerstone Science Laboratory, Center for Genome Editing, Laboratory of Advanced Breeding Technologies, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: