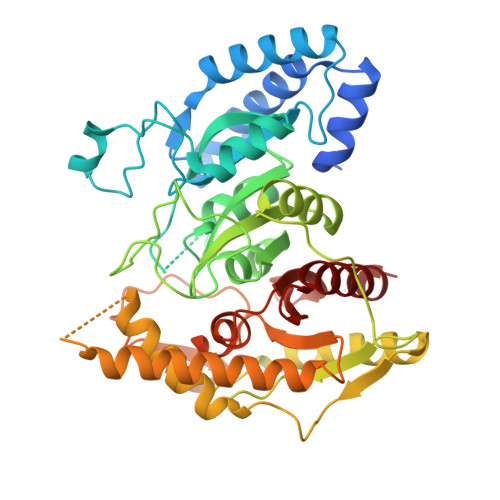
Discovery and Engineering of a Urethanase for Enhanced Depolymerization of Polyurethane
Li, Z., Fan, S., Chen, Y., Xia, W., Zhou, H., Zhou, X., Qin, B., Qin, H., Dong, W., Gu, Q., Zhu, H., Bornscheuer, U.T., Wei, R., Han, X., Liu, W.(2025) ACS Catal 15: 10768-10779