Structural basis of herpes simplex type 1 virus DNA polymerase holoenzyme in DNA replication and acyclovir inhibition and proof-reading
Wu, Y.Q., Chen, X.L., Jiang, Z.Y., Li, D.Y., Zhang, Z.Y., Dong, C.J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA polymerase | 1,235 | Human alphaherpesvirus 1 strain 17 | Mutation(s): 0 Gene Names: UL30 EC: 2.7.7.7 (PDB Primary Data), 3.1.26.4 (UniProt) | 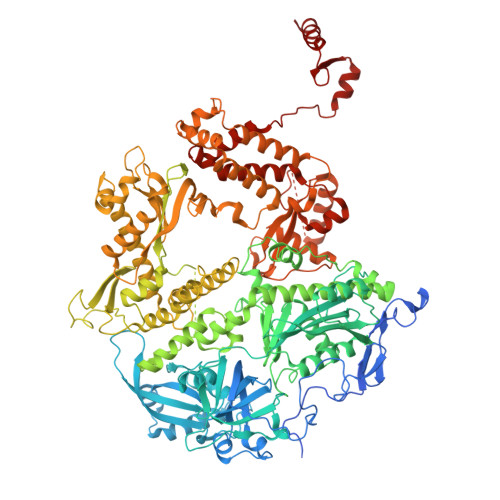 | |
UniProt | |||||
Find proteins for P04293 (Human herpesvirus 1 (strain 17)) Explore P04293 Go to UniProtKB: P04293 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04293 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA polymerase processivity factor | 513 | Human alphaherpesvirus 1 strain 17 | Mutation(s): 0 Gene Names: UL42 | 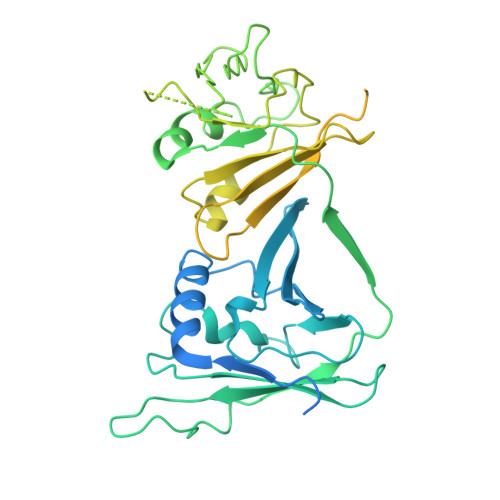 | |
UniProt | |||||
Find proteins for P10226 (Human herpesvirus 1 (strain 17)) Explore P10226 Go to UniProtKB: P10226 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P10226 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (27-MER) | C [auth T] | 27 | synthetic construct | 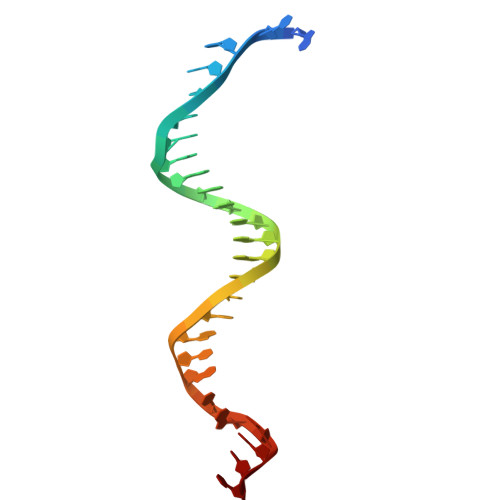 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*AP*GP*GP*CP*CP*AP*TP*AP*CP*GP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3') | D [auth P] | 23 | synthetic construct |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AVP (Subject of Investigation/LOI) Query on AVP | H [auth T] | ACYCLOVIR TRIPHOSPHATE C8 H14 N5 O12 P3 AYWLSIKEOSXJLA-UHFFFAOYSA-N |  | ||
| CA Query on CA | E [auth A], F [auth A], G [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32250710142 |