Structures and mechanisms of serine protease inhibitors of Trichinella spiralis and Trichinella pseudospiralis
Chen, C., Xue, L.R.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Serine protease inhibitor 1 serpin | 377 | Trichinella pseudospiralis | Mutation(s): 0 | 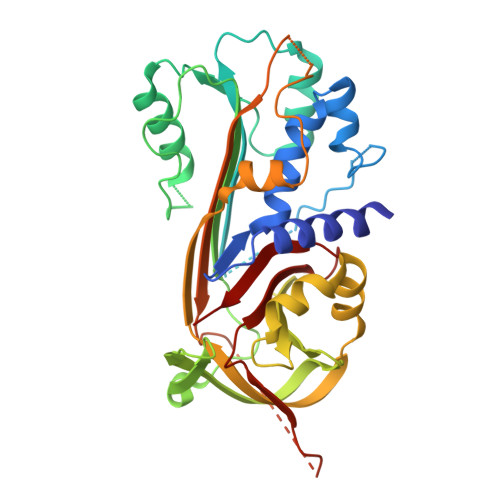 | |
UniProt | |||||
Find proteins for A0A0V1JI71 (Trichinella pseudospiralis) Explore A0A0V1JI71 Go to UniProtKB: A0A0V1JI71 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0V1JI71 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 175.498 | α = 90 |
| b = 175.498 | β = 90 |
| c = 63.425 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |