Structures of Multiple Peptide Resistance Factor from Pseudomonas aeruginosa
Jha, S., Vinothkumar, K.R.To be published.
Experimental Data Snapshot
Starting Models: experimental, in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Phosphatidylglycerol lysyltransferase | 903 | Pseudomonas aeruginosa PAO1 | Mutation(s): 0 Gene Names: PA0920 EC: 2.3.2.3 Membrane Entity: Yes | 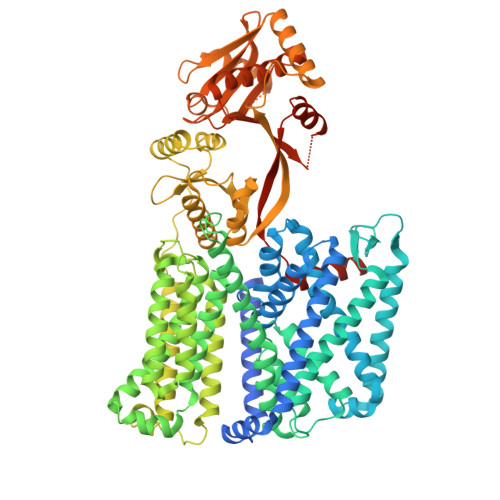 | |
UniProt | |||||
Find proteins for Q9I537 (Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1)) Explore Q9I537 Go to UniProtKB: Q9I537 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9I537 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| J4U Query on J4U | C [auth A], W [auth B] | (2~{R},3~{S},4~{S},5~{S},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{S},4~{R},5~{R},6~{R})-2-(hydroxymethyl)-6-[2-[[(2~{R},3~{S},4~{R},5~{R},6~{S})-6-(hydroxymethyl)-5-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]oxymethyl]-4-[(1~{R},2~{R},4~{S},5'~{R},6~{R},7~{R},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxy-butoxy]-4,5-bis(oxidanyl)oxan-3-yl]oxy-oxane-3,4,5-triol C56 H92 O25 LKBFXDNKNZXHHW-RFRGGUJUSA-N |  | ||
| PGT Query on PGT | CA [auth B] D [auth A] DA [auth B] E [auth A] EA [auth B] | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE C40 H79 O10 P KBPVYRBBONZJHF-AMAPPZPBSA-N |  | ||
| PTY Query on PTY | AA [auth B] BA [auth B] G [auth A] H [auth A] IA [auth B] | PHOSPHATIDYLETHANOLAMINE C40 H80 N O8 P NJGIRBISCGPRPF-KXQOOQHDSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.20.1-4487 |
| MODEL REFINEMENT | REFMAC | 5.8.0411 |
| RECONSTRUCTION | cryoSPARC | 4.3 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other government | Department of Atomic Energy, Government of India, RTI 4006 | |
| Department of Biotechnology (DBT, India) | India | DBT/PR12422/MED/31/287/2014 |