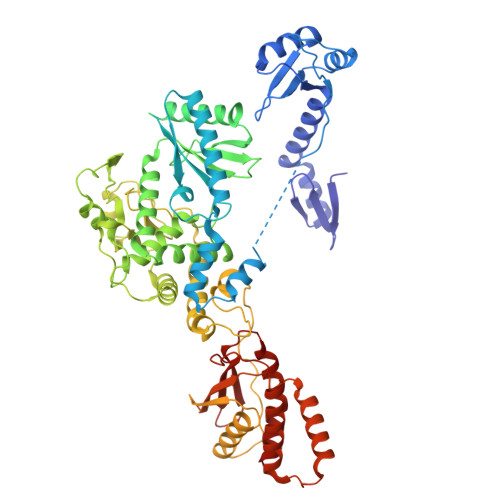
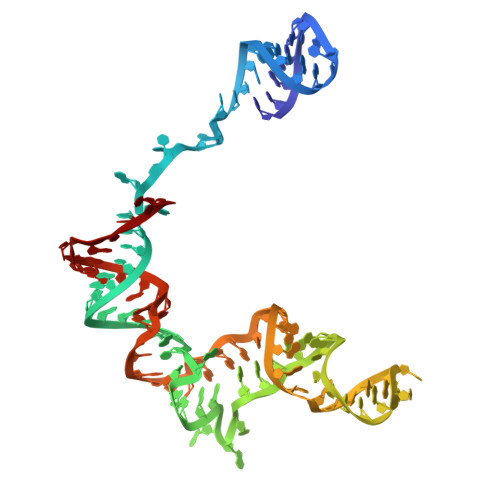
Cryo-EM structure of human TUT1:U6 snRNA complex.
Yamashita, S., Tomita, K.(2025) Nucleic Acids Res 53
- PubMed: 39831302
- DOI: https://doi.org/10.1093/nar/gkae1314
- Primary Citation of Related Structures:
9J8P - PubMed Abstract:
U6 snRNA (small nuclear ribonucleic acid) is a ribozyme that catalyzes pre-messenger RNA (pre-mRNA) splicing and undergoes epitranscriptomic modifications. After transcription, the 3'-end of U6 snRNA is oligo-uridylylated by the multi-domain terminal uridylyltransferase (TUTase), TUT1. The 3'- oligo-uridylylated tail of U6 snRNA is crucial for U4/U6 di-snRNP (small nuclear ribonucleoprotein) formation and pre-mRNA splicing. Here, we present the cryo-electron microscopy structure of the human TUT1:U6 snRNA complex. The AUA-rich motif between the 5'-short stem-loop and the telestem of U6 snRNA is clamped by the N-terminal zinc finger (ZF)-RNA recognition motif and the catalytic Palm of TUT1, and the telestem is gripped by the N-terminal ZF and the Fingers, positioning the 3'-end of the telestem in the catalytic pocket. The internal stem-loop in the 3'-stem-loop of U6 snRNA is anchored by the C-terminal kinase-associated 1 domain, preventing U6 snRNA from dislodging on the TUT1 surface during oligo-uridylylation. TUT1 recognizes the sequence and structural features of U6 snRNA, and holds the entire U6 snRNA body using multiple domains to ensure oligo-uridylylation. This highlights the specificity of TUT1 as a U6 snRNA-targeting TUTase.
- Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, 5-1-5, Kashiwanoha, Kashiwa, Chiba 277-8562, Japan.
Organizational Affiliation: