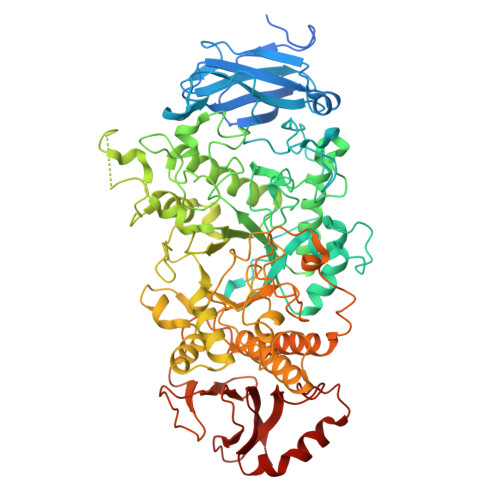
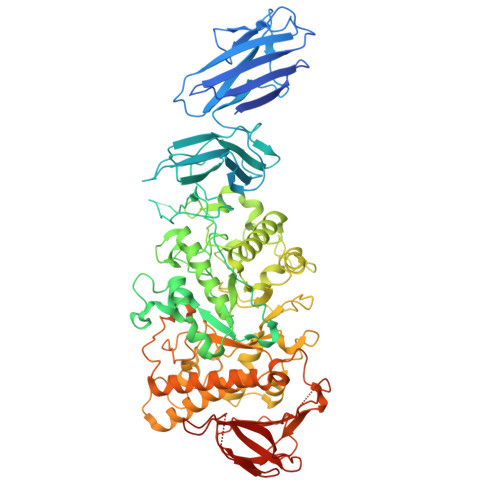
Amylopectin branch trimming and biosynthesis elucidated by the rice isoamylase ISA1-ISA2 heterocomplex.
Fan, R., Guan, Z., Zhou, G., Yang, X., Zhang, F., Wu, M., Wang, X., Liu, J., Chen, P., Liu, Y., Zhang, D., Yin, P., Yan, J.(2025) Nat Commun 16: 5638-5638
- PubMed: 40595605
- DOI: https://doi.org/10.1038/s41467-025-60944-6
- Primary Citation of Related Structures:
9J60, 9J6X, 9LFN - PubMed Abstract:
Amylopectin, the primary form of starch in plant leaves, seeds and tubers, features a tree-like architecture with branched glucose chains. Excess branches result in the formation of soluble phytoglycogen instead of starch granules. In higher plants and green algae, the debranching enzyme isoamylase ISA1 forms either homomultimer or hetero-multimer with ISA2 to facilitate branch trimming and starch granule formation, but the molecular basis remains largely unknown. In this study, we reconstitute the rice OsISA1-ISA2 complex in vitro and determine the cryo-EM structures of the OsISA1 homodimer, as well as the malto-oligosaccharide (MOS)-free and MOS-bound OsISA1-ISA2 heterocomplex. The OsISA1 dimer shows a tail-to-tail rod-like architecture, whereas the OsISA1-ISA2 complex mainly exhibits as a trimer, with OsISA2 flanking on the N-terminal segments of the dimeric OsISA1. Combined with comprehensive biochemical analyses, these structural data elucidate the organization of the ISA1-ISA2 heterocomplex in higher plants and demonstrate how ISA1 and ISA2 cooperate during amylopectin biosynthesis.
- National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan, China.
Organizational Affiliation: