Structure of the wild-type AZG2 in Arabidopsis thaliana in the trans-Zeatin-bound state-2 at pH 7.4
Sun, L., Liu, X., Wei, H., Yang, Z., Ying, W.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Adenine/guanine permease AZG2 | 530 | Arabidopsis thaliana | Mutation(s): 0 Gene Names: AZG2, At5g50300, K6A12.16 Membrane Entity: Yes | 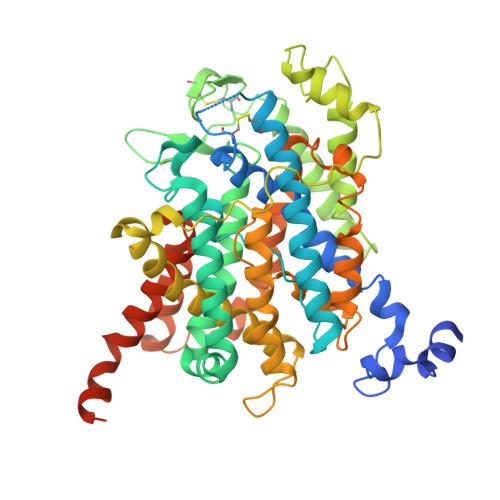 | |
UniProt | |||||
Find proteins for Q84MA8 (Arabidopsis thaliana) Explore Q84MA8 Go to UniProtKB: Q84MA8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q84MA8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZEA (Subject of Investigation/LOI) Query on ZEA | C [auth A], D [auth B] | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol C10 H13 N5 O UZKQTCBAMSWPJD-FARCUNLSSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.20.1_4487: |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Science Foundation (NSF, China) | China | 32322041, 32321001, 31900885 |