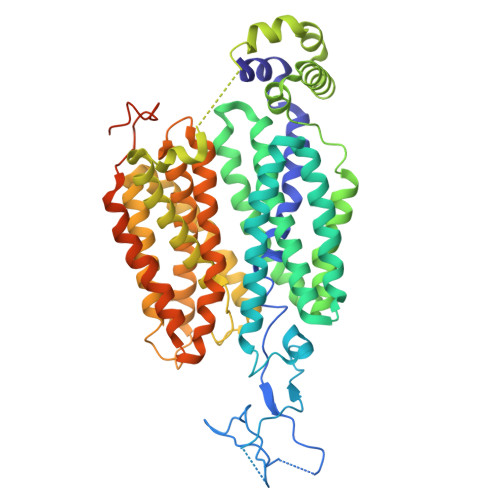
Structures and membrane interactions of human OAT1 in complex with clinical used drugs.
Wu, X., Luo, Y., Feng, S., Ma, H., Ke, B., Wang, K., Su, Z., Yang, D.(2025) Sci Adv 11: eads5405-eads5405
- PubMed: 39951534
- DOI: https://doi.org/10.1126/sciadv.ads5405
- Primary Citation of Related Structures:
9J02, 9J04, 9J06 - PubMed Abstract:
Organic anion transporters (OATs) in mammals mediate the renal excretion of numerous structurally diverse organic anionic compounds. Therapeutically inhibiting OATs has emerged as a strategy to modulate the elimination or retention of these substrates. Among them, OAT1 plays a pivotal role in the pharmacokinetics and drug-drug interactions of a wide range of prescription medications. Despite extensive structural investigations, the molecular structure, and basis of polyspecific anionic drug recognition of human OAT1 (hOAT1) have remained elusive. Here, we present cryogenic electron microscopy structures of hOAT1 and its complexes with the antiviral drug cidofovir and an FDA-approved type II diabetes medication glibenclamide, respectively. Our findings reveal that both cidofovir and glibenclamide bind to a central binding site, capturing the transporter in inward-facing conformations. These structures elucidate how specific residues within the central site orchestrate the binding of chemically diverse inhibitors and provide a structural basis for the drug recognition mechanism of hOAT1.
- Department of Urology, Institute of Urology (Laboratory of Reconstructive Urology), West China Hospital, Sichuan University, Chengdu, Sichuan 610044, China.
Organizational Affiliation: