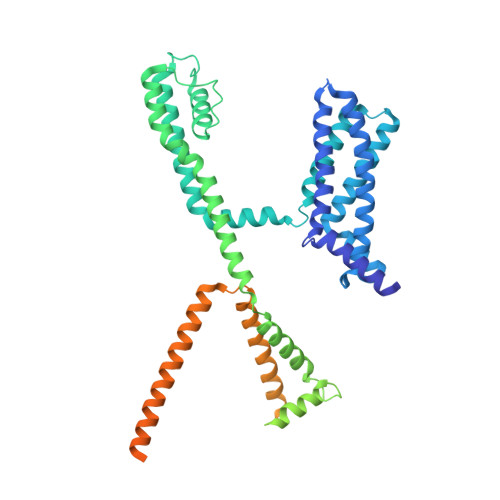
Small molecule inhibits KCNQ channels with a non-blocking mechanism.
Li, J., Yang, Z., Zhang, S., Ye, Y., He, J., Zhang, Y., Han, H., Kong, W., Liu, J., Min, Y., Shen, J., Mei, L., Chen, Z., Hou, P., Guo, J., Zhang, Q., Yang, H.(2025) Nat Chem Biol 21: 1100-1109
- PubMed: 39814994
- DOI: https://doi.org/10.1038/s41589-024-01834-8
- Primary Citation of Related Structures:
9IXY, 9IXZ - PubMed Abstract:
Voltage-gated ion channels (VGICs) are crucial targets for neuropsychiatric therapeutics owing to their role in controlling neuronal excitability and the established link between their dysfunction and neurological diseases, highlighting the importance of identifying modulators with distinct mechanisms. Here we report two small-molecule modulators with the same chemical scaffold, Ebio2 and Ebio3, targeting a potassium channel KCNQ2, with opposite effects: Ebio2 acts as a potent activator, whereas Ebio3 serves as a potent and selective inhibitor. Guided by cryogenic electron microscopy, patch-clamp recordings and molecular dynamics simulations, we reveal that Ebio3 attaches to the outside of the inner gate, employing a unique non-blocking inhibitory mechanism that directly squeezes the S6 pore helix to inactivate the KCNQ2 channel. Ebio3 also showed efficacy in inhibiting currents of KCNQ2 pathogenic gain-of-function mutations, presenting an avenue for VGIC-targeted therapies. Overall, these findings contribute to the understanding of KCNQ2 inhibition and provide insights into developing selective, non-blocking VGIC inhibitors.
- Shanghai Key Laboratory of Regulatory Biology, Institute of Biomedical Sciences and School of Life Sciences, East China Normal University, Shanghai, China.
Organizational Affiliation: