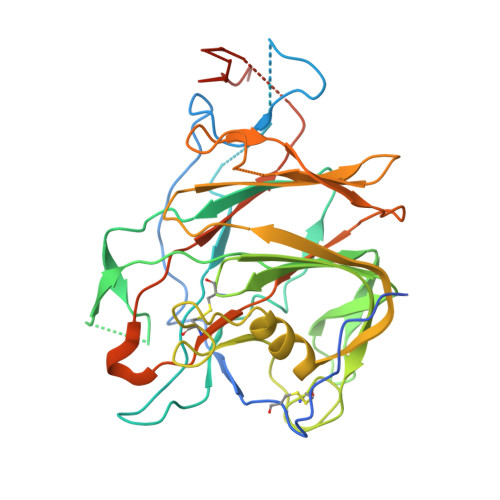
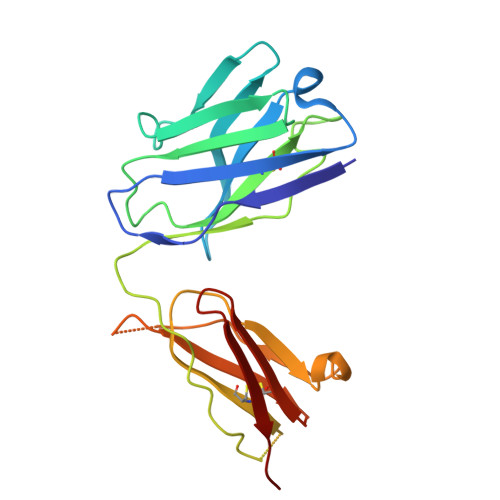
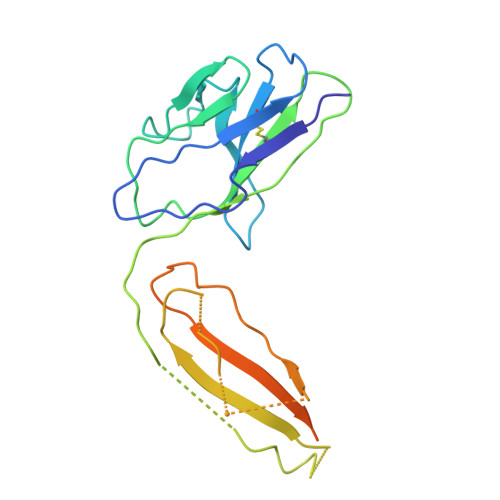
A Novel Antibody Against the Non-Receptor-Binding Domain Region of Middle East Respiratory Syndrome Coronavirus Spike Protein.
Lee, S.Y., Woo, H.M., Jeon, H., Kim, N., Kim, D.S., Park, C.K., Kim, H.J., Kim, K.C., Lee, J.Y., Park, K., Yoo, Y., Choi, K., Lee, H.(2025) J Infect Dis 232: 982-992
- PubMed: 40241666
- DOI: https://doi.org/10.1093/infdis/jiaf202
- Primary Citation of Related Structures:
9IXV - PubMed Abstract:
The Middle East respiratory syndrome coronavirus (MERS-CoV) was first identified in 2012 and has since spread worldwide. To date, no vaccines or therapeutics against MERS have been approved for clinical use. The spike (S) protein of MERS-CoV facilitates attachment and fusion with target cell membranes. Therefore, inhibiting S protein attachment represents a key therapeutic strategy for treating early MERS-CoV infection. Herein, we present seven human neutralizing antibodies (KNIH-58, -68, -72, -78, -88, -90, and -95) against MERS-CoV. KNIH-58 and -68 bound to the receptor-binding subdomain (RBD) of the spike protein, while the other five monoclonal antibodies (mAbs) did not. KNIH-88, which targets the non-RBD region, exhibited potent neutralizing activities in vitro and in a transgenic mouse model, with similar results for KNIH-58. Structural analysis of KNIH-88 bound to the spike protein revealed novel epitopes in the non-RBD region. These findings may facilitate therapeutic and prophylactic antibody development against MERS-CoV.
- Division of Emerging Virus and Vector Research, Center for Emerging Virus Research, Korea National, Institute of Health, Korea Disease Control and Prevention Agency, Cheongju-si, Republic of Korea.
Organizational Affiliation: