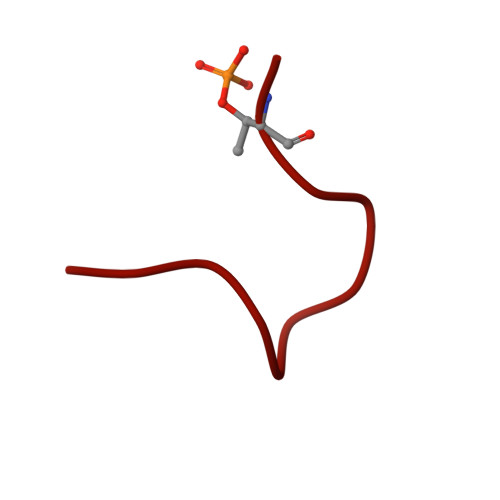
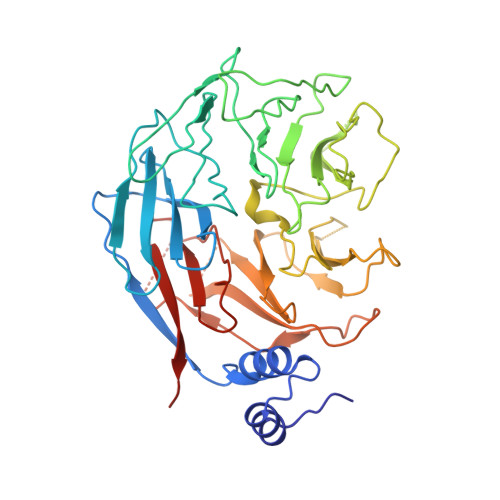
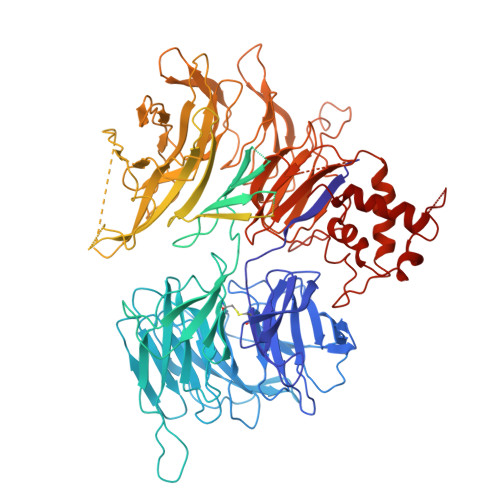
Mechanism of D-type cyclin recognition by the AMBRA1 E3 ligase receptor.
Wang, Y., Liu, M., Wang, S., Mai, X., Wang, X., Teng, F., Lyu, T., Su, M.Y., Stjepanovic, G.(2025) Sci Adv 11: eadu8708-eadu8708
- PubMed: 40408472
- DOI: https://doi.org/10.1126/sciadv.adu8708
- Primary Citation of Related Structures:
9IVD - PubMed Abstract:
AMBRA1 is a tumor suppressor protein that functions as a substrate receptor in the ubiquitin conjugation system and regulates the stability of D-type cyclins and cell proliferation. Here, we present the cryo-EM structure of cyclin D1-bound AMBRA1-DDB1 complex at 3.55-Å resolution. The structure reveals a substrate interaction surface on the AMBRA1 WD40 domain that specifically binds to the C-terminal region of D-type cyclins. This interaction is dependent on the phosphorylation of Thr 286 residue in the C-terminal phosphodegron site of D-type cyclins. The phosphodegron motif folds into a turn-like conformation, followed by a 3 10 helix that promotes its assembly with AMBRA1. In addition, we show that AMBRA1 mutants, which are defective in cyclin D1 binding, lead to cyclin D1 accumulation and DNA damage. Understanding the AMBRA1-D-type cyclin structure enhances the knowledge of the molecular mechanisms that govern the cell cycle control and may lead to potential therapeutic approaches for cancers linked to abnormal cyclin D activity.
- Kobilka Institute of Innovative Drug Discovery, School of Medicine, The Chinese University of Hong Kong, Shenzhen, Guangdong 518172, China.
Organizational Affiliation: