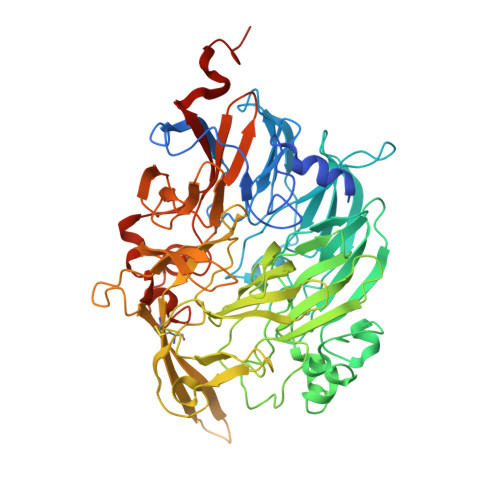
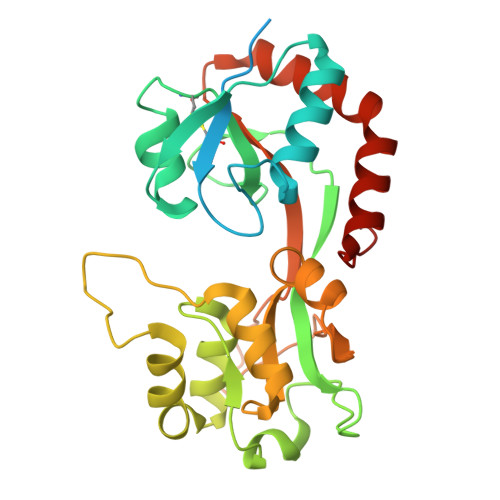
Deciphering the assembly process of PQQ dependent methanol dehydrogenase.
Zhou, H., Sun, J., Cheng, J., Wu, M., Bai, J., Li, Q., Shen, J., Han, M., Yang, C., Li, L., Liu, Y., Cao, Q., Liu, W., Xiao, H., Dong, H., Gao, F., Jiang, H.(2025) Nat Commun 16: 6672-6672
- PubMed: 40683858
- DOI: https://doi.org/10.1038/s41467-025-61958-w
- Primary Citation of Related Structures:
9ISM, 9ISO - PubMed Abstract:
Pyrroloquinoline quinone (PQQ)-dependent methanol dehydrogenases (MDHs), the periplasmic metalloenzymes in Gram-negative methylotrophic bacteria, play a pivotal role in methane and methanol bio-utilization. Although the structures of many PQQ-dependent MDHs have been resolved, including the canonical heterotetrameric enzymes composed of two MxaF and two MxaI subunits with a molecule of PQQ and a calcium ion in the active site in MxaF, the biogenesis of these enzymes remains elusive. Here, we characterize a chaperone, MxaJ, responsible for PQQ incorporation by reconstructing a PQQ-dependent MDH assembly system in Escherichia coli. Using cryo-electron microscopy, we capture the structures of the intermediate complexes formed by the chaperone MxaJ and catalytic subunit MxaF during PQQ-dependent MDH maturation, revealing a chaperone-mediated molecular mechanism of cofactor incorporation. These findings not only advance our understanding on the biogenesis of PQQ-dependent MDH, but also provide an alternative engineering way for methane and methanol bioconversion.
- State Key Laboratory of Engineering Biology for Low-Carbon Manufacturing, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, China.
Organizational Affiliation: