Establish a Least-Square Circle Feature Extraction method for identifying new DNA mimic proteins
Liao, B.C., Chen, Y.H., Chou, H.H., Hsu, K.C., Ko, T.P., Liu, C.Y., Lin, S.J., Chen, Y.C., Hsieh, S.Y., Wang, H.C.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Uracil-DNA glycosylase | 226 | Staphylococcus aureus | Mutation(s): 0 Gene Names: ung, SAR0586 EC: 3.2.2.27 | 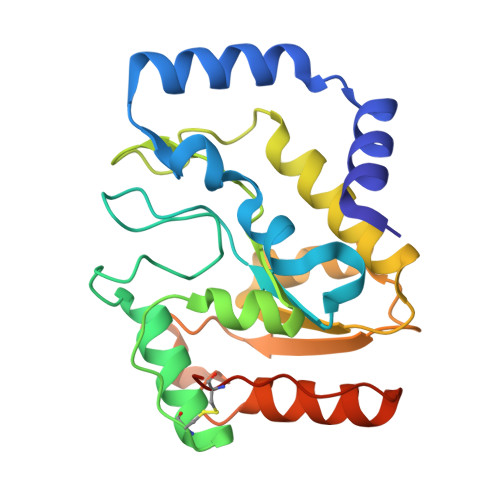 | |
UniProt | |||||
Find proteins for Q2G0J7 (Staphylococcus aureus (strain NCTC 8325 / PS 47)) Explore Q2G0J7 Go to UniProtKB: Q2G0J7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2G0J7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Staphylococcus epidermidis uracil-DNA glycosylase inhibitor (SeUGI) | 132 | Staphylococcus epidermidis | Mutation(s): 0 | 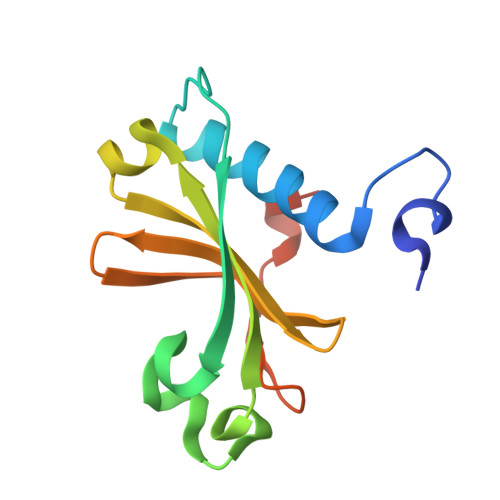 | |
UniProt | |||||
Find proteins for A0A141HMG4 (Staphylococcus aureus subsp. aureus RN4220) Explore A0A141HMG4 Go to UniProtKB: A0A141HMG4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A141HMG4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 (Subject of Investigation/LOI) Query on SO4 | C [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 60.895 | α = 90 |
| b = 75.86 | β = 90 |
| c = 77.056 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data scaling |
| MOLREP | phasing |
| BUCCANEER | model building |
| PHENIX | refinement |
| HKL-2000 | data processing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministry of Science and Technology (MoST, Taiwan) | Taiwan | NSTC 112-2628-B-038 -011 |