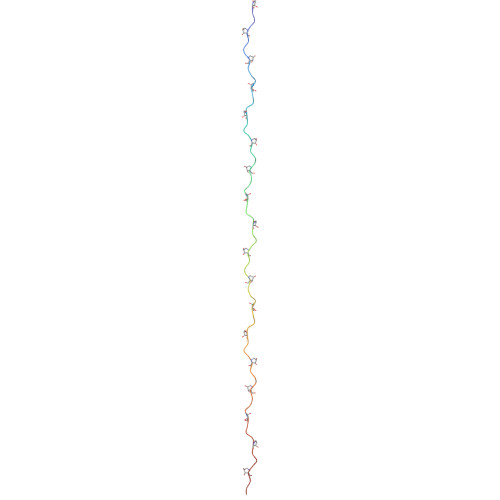CryoSeek II: Cryo-EM analysis of glycofibrils from freshwater reveals well-structured glycans coating linear tetrapeptide repeats.
Wang, T., Huang, W., Xu, K., Sun, Y., Zhang, Q.C., Yan, C., Li, Z., Yan, N.(2025) Proc Natl Acad Sci U S A 122: e2423943122-e2423943122
- PubMed: 39739783
- DOI: https://doi.org/10.1073/pnas.2423943122
- Primary Citation of Related Structures:
9IKM - PubMed Abstract:
Despite the recent breakthrough in structure determination and prediction of proteins, the structural investigation of carbohydrates remains a challenge. Here, we report the cryo-EM analysis of a glycofibril found in the freshwater in the Tsinghua Lotus Pond. The fibril, which we name TLP-4, is made of a linear chain of tetrapeptide repeats coated with >4 nm thick glycans. In each repeat, two glycans are O-linked to a 3,4-dihydroxyproline and another glycan attaches to the adjacent Ser or Thr. The fibril structure is entirely maintained through glycan packing. Bioinformatic analysis confirms the conservation of the TLP-4 repeats across species, suggesting the existence of a large number of glycofibrils to be discovered. Our findings not only provide valuable insights into the structural roles of glycans in bio-assemblies but also demonstrate the potential of our recently formulated research strategy of CryoSeek to find bioentities and establish prototypes for structural studies of carbohydrates.
- Beijing Frontier Research Center for Biological Structures, State Key Laboratory of Membrane Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: