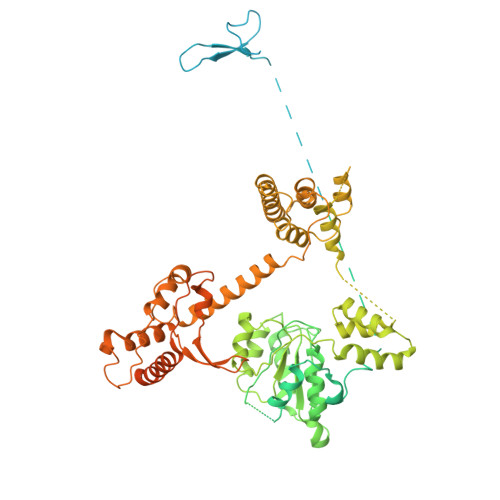
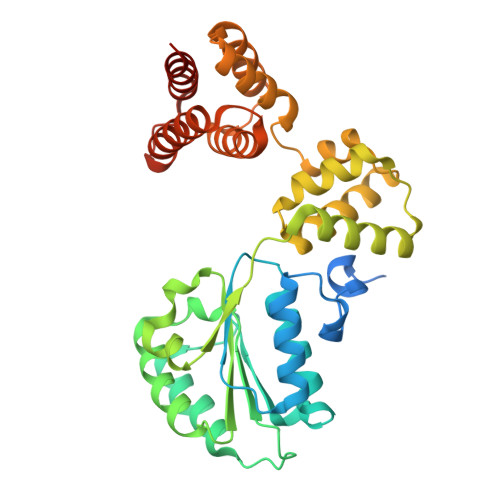
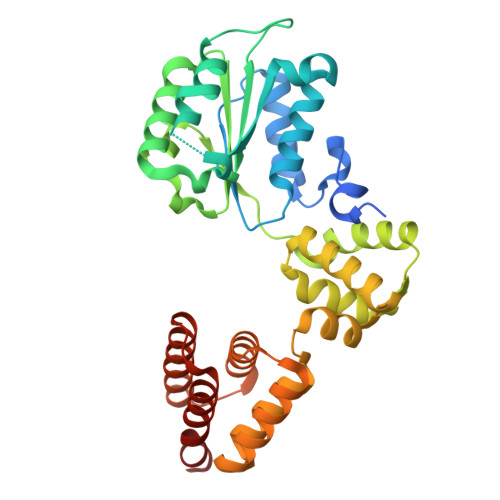
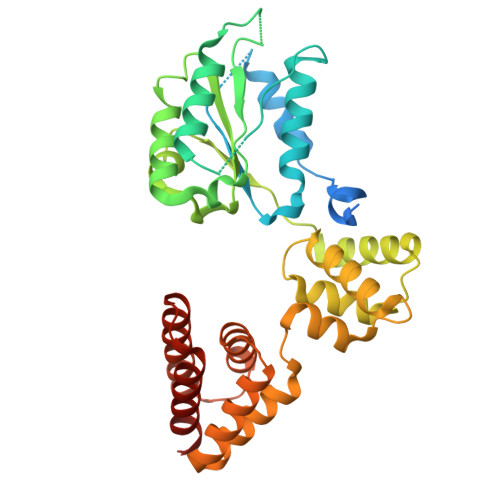
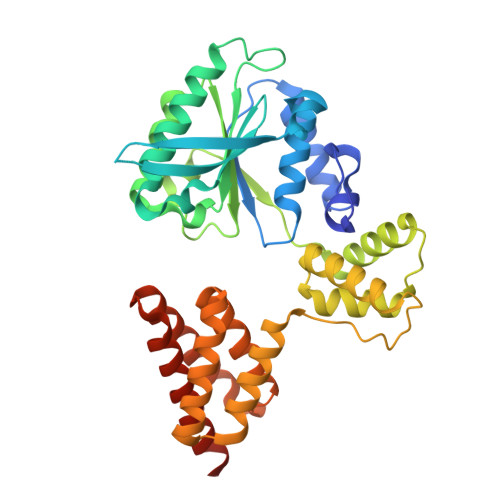
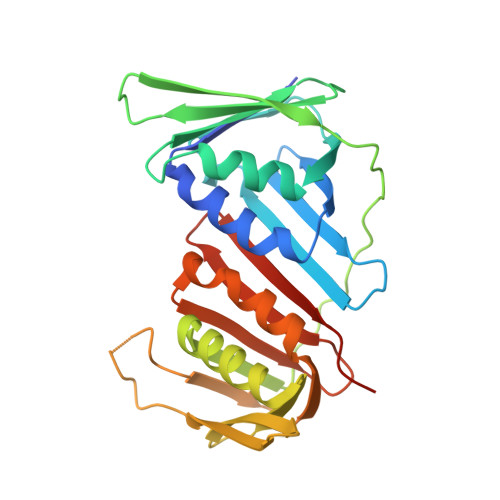
Structure of the human CTF18-RFC clamp loader bound to PCNA.
Briola, G.R., Tehseen, M., Al-Amodi, A., Young, G., Danazumi, A.U., Nguyen, P.Q., Savva, C.G., Hedglin, M., Hamdan, S.M., Biasio, A.(2025) bioRxiv
- PubMed: 40777363
- DOI: https://doi.org/10.1101/2024.05.08.593111
- Primary Citation of Related Structures:
8ZWO, 9IIN - PubMed Abstract:
Sliding clamps like PCNA are crucial processivity factors for replicative polymerases, requiring specific clamp loaders for loading onto DNA. The human alternative clamp loader CTF18-RFC interacts with the leading strand polymerase Pol ε and loads PCNA onto primer/template DNA using its RFC pentameric module. Here, we provide a structural characterization of the human CTF18-RFC complex and its interaction with PCNA. Our cryo-EM data support that the Ctf8 and Dcc1 subunits of CTF18-RFC, which form the regulatory module interacting with Pol ε, are flexibly tethered to the RFC module. A 2.9 Å cryo-EM structure shows the RFC module bound to PCNA in an auto-inhibited conformation similar to the canonical RFC loader, marking the initial step of the clamp-loading reaction. The unique RFC1 (Ctf18) large subunit of CTF18-RFC, which based on the cryo-EM map shows high relative flexibility, is anchored to PCNA through an atypical low-affinity PIP box in the AAA+ domain and engages the RFC5 subunit using a novel β-hairpin at the disordered N-terminus. We show that deletion of this β-hairpin impairs the CTF18-RFC-PCNA complex stability, slows down clamp loading, and decreases the rate of primer synthesis by Pol ε. Our research identifies distinctive structural characteristics of the human CTF18-RFC complex, providing insights into its role in PCNA loading and the stimulation of leading strand synthesis by Pol ε.
- Bioscience Program, Division of Biological and Environmental Sciences and Engineering, King Abdullah University of Science and Technology, Thuwal 23955, Saudi Arabia.
Organizational Affiliation: