Structural and mechanistic insights into the sequential dsDNA cleavage by SpCas12f1.
Madariaga-Marcos, J., Baltramonaitis, M., Henkel-Heinecke, S., Kauert, D.J., Irmisch, P., Bigelyte-Stankeviciene, G., Silanskas, A., Karvelis, T., Siksnys, V., Sasnauskas, G., Seidel, R.(2025) Nucleic Acids Res 53
- PubMed: 40650969
- DOI: https://doi.org/10.1093/nar/gkaf588
- Primary Citation of Related Structures:
9I8Y - PubMed Abstract:
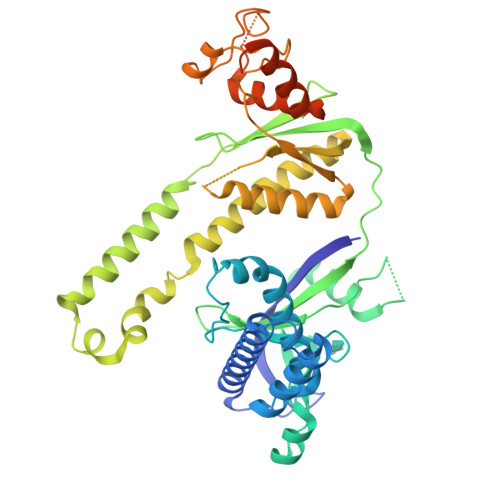
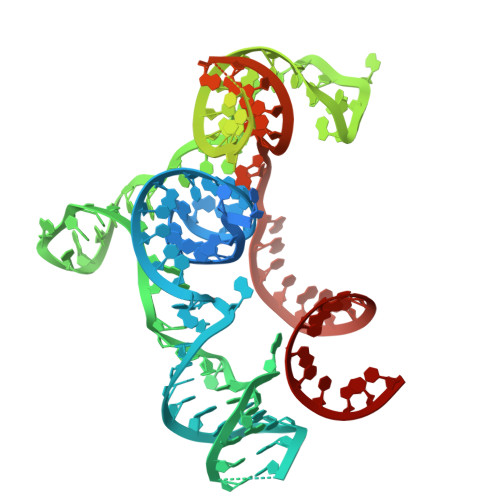
Miniature CRISPR-Cas12f1 effector complexes have recently attracted considerable interest for genome engineering applications due to their compact size. Unlike other Class 2 effectors, Cas12f1 functions as a homodimer bound to a single ∼200 nt RNA. While the basic biochemical properties of Cas12f1, such as its use of a single catalytic center for catalysis, have been characterized, the orchestration of the different events occurring during Cas12f1 reactions remained little explored. To gain insights into the dynamics and mechanisms involved in DNA recognition and cleavage by Cas12f1 from Syntrophomonas palmitatica (SpCas12f1), we solved the structure of SpCas12f1 bound to target DNA and employed single-molecule magnetic tweezers measurements in combination with ensemble kinetic measurements. Our data indicate that SpCas12f1 forms 18 bp R-loops, in which local contacts of the protein to the R-loop stabilize R-loop intermediates. DNA cleavage is catalyzed by a single SpCas12f1 catalytic center, which first rapidly degrades a ∼11 bp region on the nontarget strand by cutting at random sites. Subsequent target strand cleavage is slower and requires at least a nick in the nontarget strand.
- Peter Debye Institute for Soft Matter Physics, Universität Leipzig, Leipzig 04103, Germany.
Organizational Affiliation: