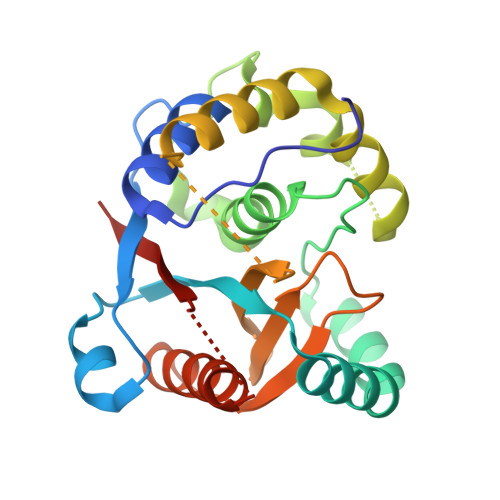
Structure and activity of the essential UCH family deubiquitinase DUB16 from Leishmania donovani.
Brannigan, J.A., Kamran, M., Jones, N.G., Nightingale, E.M., Dodson, E.J., Ejazi, S.A., Mottram, J.C., Ali, N., Wilkinson, A.J.(2025) Biochem J 482: 969-988
- PubMed: 40570172
- DOI: https://doi.org/10.1042/BCJ20253107
- Primary Citation of Related Structures:
9I77 - PubMed Abstract:
In Leishmania parasites, as for their hosts, the ubiquitin proteasome system is important for cell viability. As part of a systematic gene deletion study, it was discovered that four cysteine protease type deubiquitinases (DUBs) are essential for parasite survival in the promastigote stage, including DUB16. Here we have purified and characterised recombinant DUB16 from Leishmania donovani, which belongs to the ubiquitin C-terminal hydrolase (UCH) family. DUB16 efficiently hydrolyses C-terminal aminocoumarin and rhodamine conjugates of ubiquitin consistent with proposed cellular roles of UCH-type DUBs in regenerating free monomeric ubiquitin from small molecule ubiquitin adducts arising from adventitious metabolic processes. The crystal structure of DUB16 reveals a typical UCH-type deubiquitinase fold, and a relatively short and disordered crossover loop that appears to restrict access to the catalytic cysteine. At close to stoichiometric enzyme to substrate ratios, DUB16 exhibits deubiquitinase activity towards diubiquitins linked through isopeptide bonds between Lys11, Lys48 or Lys63 residues of the proximal ubiquitin and the C-terminus of the distal ubiquitin. With 100-1000-fold higher turnover rates, DUB16 cleaves the ubiquitin-ribosomal L40 fusion protein to give the mature products. A DUB-targeting cysteine-reactive cyanopyrrolidine compound, IMP-1710, inhibits DUB16 activity. IMP-1710 was shown in promastigote cell viability assays to have parasite killing activity with EC50 values of 1-2 M, comparable to the anti-leishmanial drug, miltefosine. L. mexicana parasites engineered to overproduce DUB16 showed a modest increase in resistance to IMP-1710, providing evidence that IMP-1710 inhibits DUB16 in vivo. While it is highly likely that IMP-1710 has additional targets, these results suggest that DUB16 is a potential target for the development of new anti-leishmanial compounds.
- York Structural Biology Laboratory, York Biomedical Research Institute and Department of Chemistry, University of York, York, YO10 5DD, U.K.
Organizational Affiliation: