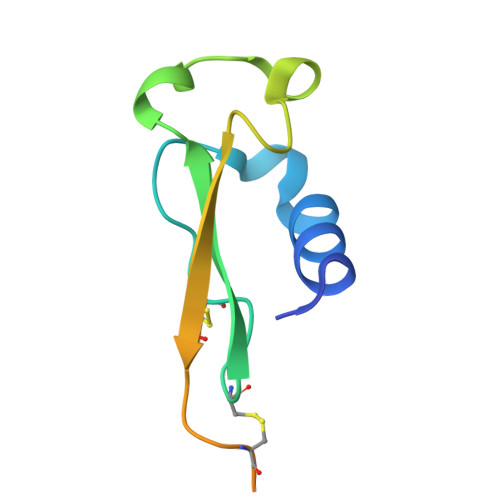
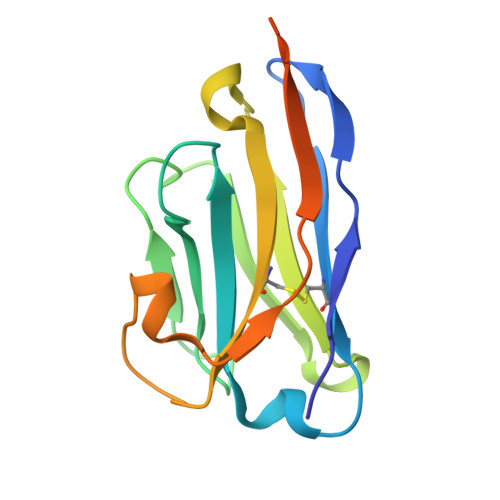
Evidence for a model of conformational change by the Plasmodium falciparum circumsporozoite protein during sporozoite development in the mosquito host through the use of camelid single-domain antibodies.
Geens, R., De Vocht, L., Aguirre-Botero, M.C., Vincke, C., Romao, E., Magez, S., Muyldermans, S., Amino, R., Sterckx, Y.G.-.J.(2025) Infect Immun 93: e0008125-e0008125
- PubMed: 40293231
- DOI: https://doi.org/10.1128/iai.00081-25
- Primary Citation of Related Structures:
9HZJ, 9HZK - PubMed Abstract:
Plasmodium sporozoites (SPZs) are formed in the Anopheles mosquito midgut from where they travel to the salivary glands and subsequently to the mammalian liver after deposition into the skin. The SPZ's main surface antigen, the circumsporozoite protein (CSP), plays a pivotal role in SPZ biology and constitutes the immunodominant target for host antibodies. In this study, we raised single-domain antibodies (sdAbs) against CSP from P. falciparum (PfCSP) by immunizing two alpacas with recombinant versions of the antigen. We found that all identified sdAbs specifically target PfCSP's globular [Formula: see text]TSR domain without cross-reacting with P. berghei CSP. Further characterization revealed that most sdAbs recognize native PfCSP on the SPZ surface, although they do not have any inhibitory effect on hepatocyte binding and invasion. Structural studies showed that all binders target the previously identified [Formula: see text]-epitope, confirming the non-protective nature of this epitope. Comparison of sdAb binding to midgut and salivary gland SPZs revealed a shift in the exposure and accessibility of the [Formula: see text]-epitope. Hence, our findings provide further evidence that CSP undergoes structural changes during SPZ development in the mosquito host.
- Laboratory of Medical Biochemistry (LMB) and the Infla-Med Centre of Excellence, Department of Pharmaceutical Sciences, University of Antwerp, Antwerp, Belgium.
Organizational Affiliation: