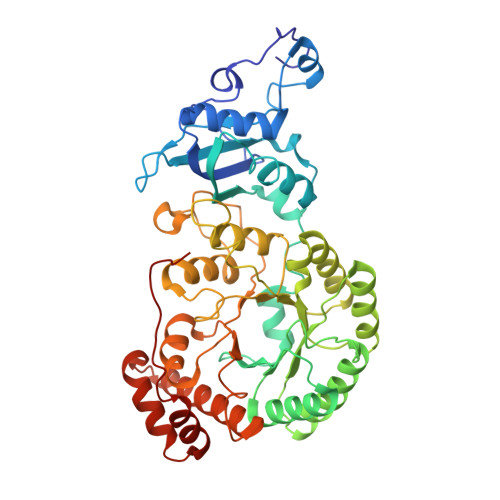
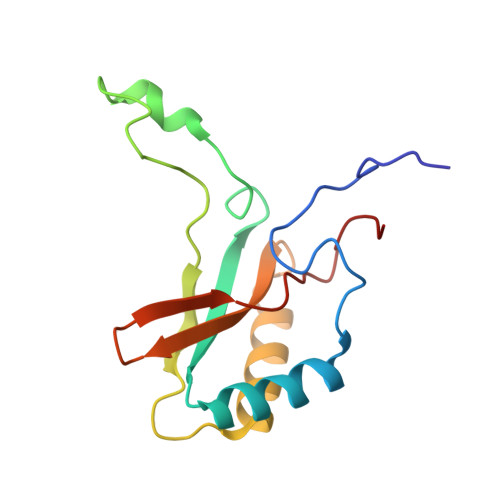
In-cell structure and variability of pyrenoid Rubisco.
Elad, N., Hou, Z., Dumoux, M., Ramezani, A., Perilla, J.R., Zhang, P.(2025) Nat Commun 16: 7763-7763
- PubMed: 40835820
- DOI: https://doi.org/10.1038/s41467-025-62998-y
- Primary Citation of Related Structures:
9HVM - PubMed Abstract:
Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) is central to global CO 2 fixation. In eukaryotic algae, its catalytic efficiency is enhanced through the pyrenoid - a protein-dense organelle within the chloroplast that concentrates CO 2 . Although Rubisco structure has been extensively studied in vitro, its native structure, dynamics and interactions within the pyrenoid remain elusive. Here, we present the native Rubisco structure inside the green alga Chlamydomonas reinhardtii determined by cryo-electron tomography and subtomogram averaging of cryo-focused ion beam milled cells. Multiple structural subsets of Rubisco are identified, stochastically distributed throughout the pyrenoid. While Rubisco adopts an active conformation in the best-resolved map, comparison among the subsets reveals significant local variations at the active site, at the large subunit dimer interfaces, and at binding protein contact regions. These findings offer a comprehensive understanding of the structure, dynamics, and functional organization of native Rubisco within the pyrenoid, providing valuable insights into its critical role in CO 2 fixation.
- Department of Chemical Research Support, Weizmann Institute of Science, Rehovot, Israel.
Organizational Affiliation: