Structural and biochemical characterization of yeast Tcd enzymes installing the epigenetic modification ct6A in tRNA
Hirschmann, J., Sonntag, R., Heiss, M., Wegrzyn, E., Heinemeyer, W., Carell, T., Huber, E.M.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| tRNA threonylcarbamoyladenosine dehydratase 1 | 382 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: TCD1, YHR003C EC: 6.1 | 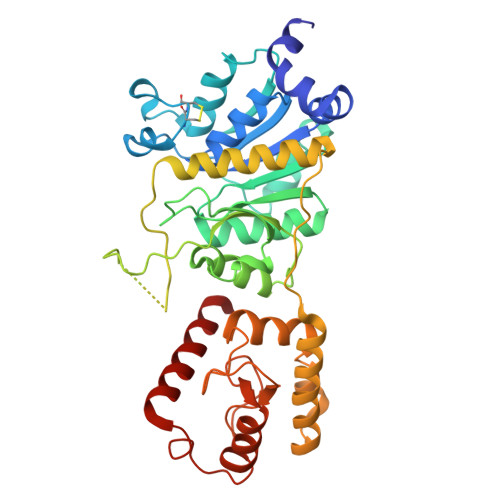 | |
UniProt | |||||
Find proteins for P38756 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P38756 Go to UniProtKB: P38756 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P38756 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AMP (Subject of Investigation/LOI) Query on AMP | E [auth A], K [auth B], P [auth C], T [auth D] | ADENOSINE MONOPHOSPHATE C10 H14 N5 O7 P UDMBCSSLTHHNCD-KQYNXXCUSA-N |  | ||
| GOL Query on GOL | L [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| CL Query on CL | F [auth A], G [auth A], Q [auth C], R [auth C], U [auth D] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | H [auth A] I [auth A] J [auth A] M [auth B] N [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 259.44 | α = 90 |
| b = 259.44 | β = 90 |
| c = 236.11 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Research Foundation (DFG) | Germany | 325871075 |