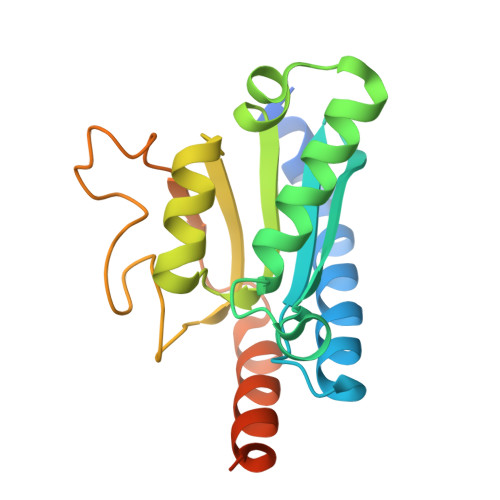
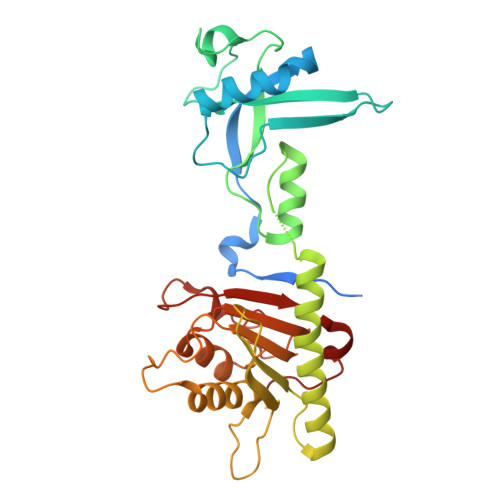
ADAT3 variants disrupt the activity of the ADAT tRNA deaminase complex and impair neuronal migration.
Del-Pozo-Rodriguez, J., Tilly, P., Lecat, R., Vaca, H.R., Mosser, L., Brivio, E., Balla, T., Gomes, M.V., Ramos-Morales, E., Schwaller, N., Salinas-Giege, T., VanNoy, G., England, E.M., Kern Lovgren, A., O'Leary, M., Chopra, M., Meave Ojeda, N., Toosi, M.B., Eslahi, A., Alerasool, M., Mojarrad, M., Pais, L.S., Yeh, R.C., Gable, D.L., Hashem, M.O., Abdulwahab, F., Rakiz Alqurashi, M., Sbeih, L.Z., Adas Blanco, O.A., Khater, R.A., Oprea, G., Rad, A., Alzaidan, H., Aldhalaan, H., Tous, E., Alsagheir, A., Alowain, M., Tamim, A., Alfayez, K., Alhashem, A., Alnuzha, A., Kamel, M., Al-Awam, B.S., Elnaggar, W., Almenabawy, N., O'Donnell-Luria, A., Neil, J.E., Gleeson, J.G., Walsh, C.A., Alkuraya, F.S., AlAbdi, L., Elkhateeb, N., Selim, L., Srivastava, S., Nedialkova, D.D., Drouard, L., Romier, C., Bayam, E., Godin, J.D.(2025) Brain 148: 3407-3421
- PubMed: 40120092
- DOI: https://doi.org/10.1093/brain/awaf109
- Primary Citation of Related Structures:
9HMM - PubMed Abstract:
The ADAT2/ADAT3 (ADAT) complex catalyzes the adenosine to inosine modification at the wobble position of eukaryotic tRNAs. Mutations in ADAT3, the catalytically inactive subunit of the ADAT2/ADAT3 complex, have been identified in patients presenting with severe neurodevelopmental disorders. Yet, the physiological function of ADAT2/ADAT3 complex during brain development remains totally unknown. Here, we investigated the role of the ADAT2/ADAT3 complex in cortical development. First, we reported 21 neurodevelopmental disorders patients carrying biallelic variants in ADAT3. Second, we used structural, biochemical, and enzymatic assays to deeply characterize the impact of those variants on ADAT2/ADAT3 structure, biochemical properties, enzymatic activity and tRNAs editing and abundance. Finally, in vivo complementation assays were performed to correlate functional deficits with neuronal migration defects in the developing mouse cortex. Our results showed that maintaining a proper level of ADAT2/ADAT3 catalytic activity is essential for radial migration of projection neurons in the developing mouse cortex. We demonstrated that the identified ADAT3 variants significantly impaired the abundance and, for some, the activity of the complex, leading to a substantial decrease in I34 levels with direct consequence on their steady-state. We correlated the severity of the migration phenotype with the degree of the loss of function caused by the variants. Altogether, our results highlight the critical role of ADAT2/ADAT3 during cortical development and provide cellular and molecular insights into the pathogenic mechanisms underlying ADAT3-related neurodevelopmental disorders.
- IGBMC, Institut de Génétique et de Biologie Moléculaire et Cellulaire, 67400 Illkirch, France.
Organizational Affiliation: