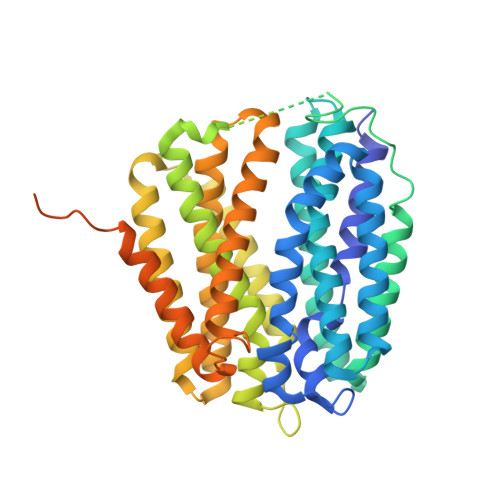
Structure-based discovery of thiamine uptake inhibitors.
Gabriel, F., Windshugel, B., Low, C.(2025) Br J Pharmacol 182: 5611-5626
- PubMed: 40702645
- DOI: https://doi.org/10.1111/bph.70133
- Primary Citation of Related Structures:
9HJD - PubMed Abstract:
Thiamine (vitamin B 1 ) is an essential coenzyme and catalyses various reactions in central metabolic pathways. Since mammals have lost the ability to synthesise thiamine de novo, this micronutrient has to be imported via the high affinity solute carriers SLC19A2 and A3 across the plasma membrane. Perturbations of these transport systems have severe effects on human health. Recent structural work on SLC19A2 and A3 have provided molecular insights into substrate and drug recognition and conformational changes during transport. Based on the analysis of the available SLC19A3 structures, we hypothesise that the binding site is rather promiscuous, allowing different small molecules to interact and potentially inhibit this transporter. We employed a computational approach, by which 538 approved and investigational drugs were docked into an ensemble of SLC19A3 cryo-EM structures, followed by experimental binding studies, transport inhibition assays, and structural validation. Eight novel compounds were identified that bind and inhibit SLC19A3. To visualise such a new drug interaction, we determined the cryo-EM structure of SLC19A3 bound to domperidone, a dopamine D 2 receptor antagonist used for the treatment of nausea and gastrointestinal disorders. Our computational work together with biochemical and cellular transport assays expands the understanding of SLC19A3-drug interactions, highlights the power of virtual screening approaches using structural ensembles, and provides a three-dimensional pharmacophore model for SLC19A3 inhibitors. These findings offer a basis for addressing drug-induced thiamine deficiencies and pre approach can be used to optimise pharmacological strategies involving SLC19A3-interacting compounds in the future.
- Centre for Structural Systems Biology (CSSB), Hamburg, Germany.
Organizational Affiliation: