Engineering and Structural Elucidation of a Sac7d-Derived IgG Fc-Specific Affitin and Its Application for the Light-Controlled Affinity Purification of Antibodies.
Veitl, F., Eichinger, A., Mayrhofer, P., Anneser, M.R., Testanera, M., Rauscher, K., Lenz, M., Skerra, A.(2025) Chembiochem 26: e202500102-e202500102
- PubMed: 40085144
- DOI: https://doi.org/10.1002/cbic.202500102
- Primary Citation of Related Structures:
9HIZ - PubMed Abstract:
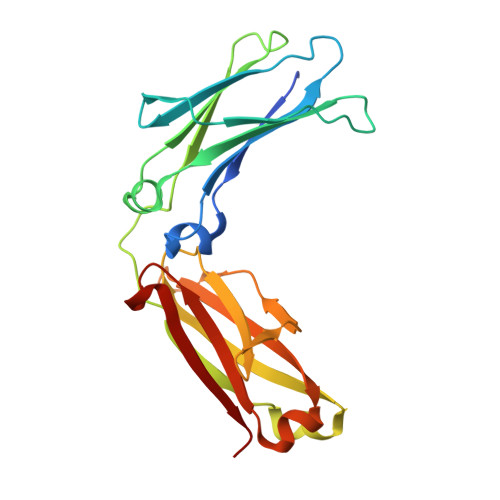
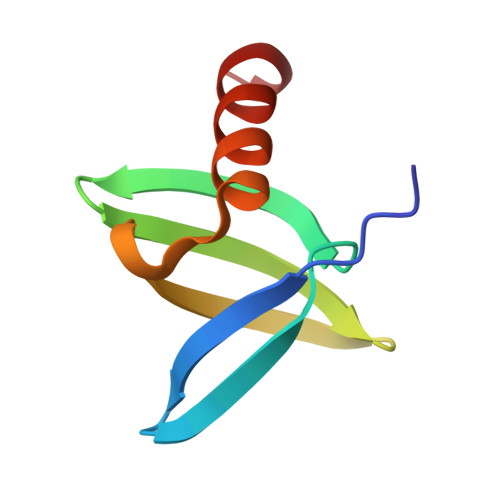
While protein A affinity chromatography is widely established for antibody purification, the acidic elution conditions often lead to protein aggregation and deamidation. Here, an alternative approach is described for the purification of antibodies utilizing an engineered binding protein based on the archaebacterial Sac7d scaffold in combination with light-controlled α-CD affinity chromatography (Excitography). Starting from a published affitin molecule, a monomeric protein version (C3A24) was engineered by substituting the unpaired thiol side chain Cys24 within the binding site by Ala, and, unexpectedly, its binding activity towards the human IgG1 Fc region was even improved (K D = 76 nM). X-ray analysis of the cocrystallized C3A24 with a recombinant human Fc fragment revealed a 2:1 stoichiometry, with a binding site at the junction between the C H 2 and C H 3 domains. Interestingly, this binding site coincides with the ones of protein A, protein G, and the neonatal Fc receptor (FcRn). The affitin/Fc interaction is dominated by a network of hydrogen bonds, whereas, unpredicted by the initial affitin design, the two C-terminal Lys residues are also involved via a salt bridge and another hydrogen bond. Using the Azo-tagged C3A24, we purified clinically relevant antibodies from cell culture medium in a single step under physiological buffer conditions.
- Chair of Biological Chemistry, School of Life Sciences, Technical University of Munich, 85354, Freising, Germany.
Organizational Affiliation: