Structural basis of serotonin reuptake inhibition by a dual mode orthosteric/allosteric antidepressant
Kalenderoglou, I.E., Nygaard, A., Vogt, C.D., Turaev, A., Pape, T., Adams, N., Newman, A.H., Loland, C.J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sodium-dependent serotonin transporter | 685 | Homo sapiens | Mutation(s): 0 Gene Names: SLC6A4, HTT, SERT Membrane Entity: Yes | 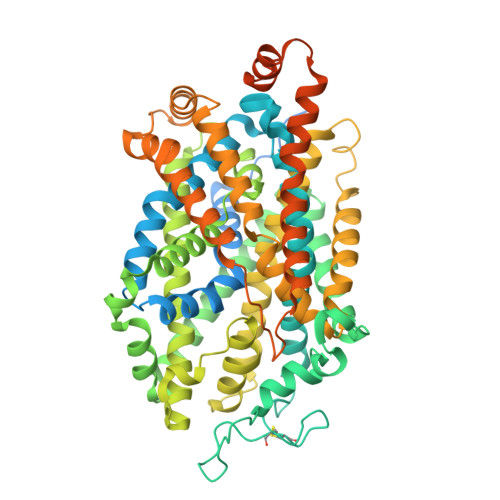 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P31645 (Homo sapiens) Explore P31645 Go to UniProtKB: P31645 | |||||
PHAROS: P31645 GTEx: ENSG00000108576 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P31645 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 15B8 Fab heavy chain | B [auth H] | 229 | Mus musculus | Mutation(s): 0 | 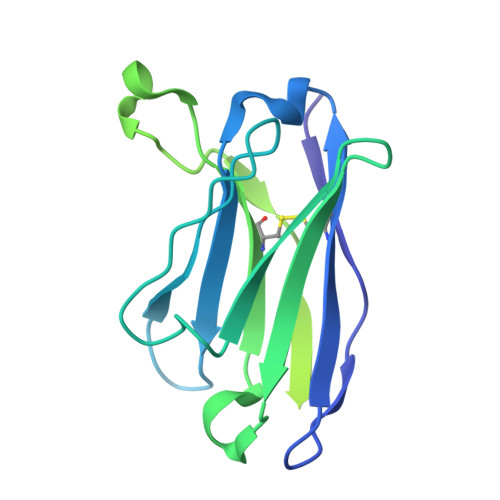 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 15B8 Fab light chain | C [auth L] | 216 | Mus musculus | Mutation(s): 0 | 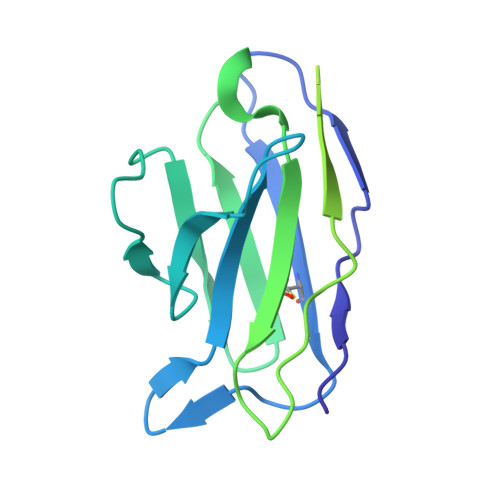 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| YG7 (Subject of Investigation/LOI) Query on YG7 | D [auth A] | 5-{4-[4-(5-cyano-1H-indol-3-yl)butyl]piperazin-1-yl}-1-benzofuran-2-carboxamide C26 H27 N5 O2 SGEGOXDYSFKCPT-UHFFFAOYSA-N |  | ||
| CL Query on CL | F [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | E [auth A], G [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Novo Nordisk Foundation | Denmark | NNF22OC0079091 |
| Independent Research Fund Denmark - Medical Sciences | Denmark | 1030-00036B |
| H2020 Marie Curie Actions of the European Commission | European Union | 860954 |
| The Carlsberg Foundation | Denmark | 1030-00036B |
| National Institutes of Health/National Institute on Drug Abuse (NIH/NIDA) | United States | Z1A DA000389 |
| Novo Nordisk Foundation | Denmark | NNF17SA0024386 |
| Novo Nordisk Foundation | Denmark | NNF22OC0075808 |