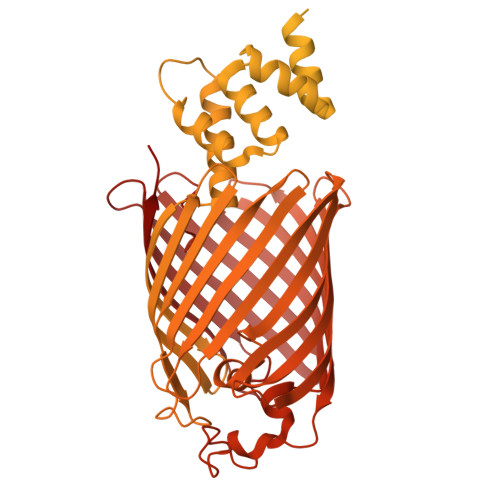
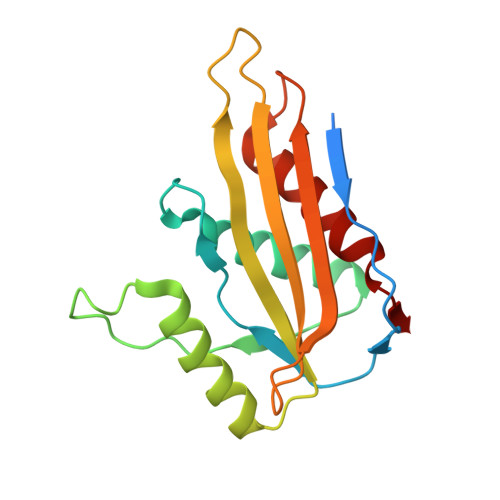
Assembly and the gating mechanism of the Pel exopolysaccharide export complex PelBC of Pseudomonas aeruginosa.
Benedens, M., Rosales-Hernandez, C., Straathof, S.A.P., Loschwitz, J., Berninghausen, O., Maglia, G., Beckmann, R., Kedrov, A.(2025) Nat Commun 16: 5249-5249
- PubMed: 40473691
- DOI: https://doi.org/10.1038/s41467-025-60605-8
- Primary Citation of Related Structures:
9H80 - PubMed Abstract:
The pathogen Pseudomonas aeruginosa enhances its virulence and antibiotic resistance upon formation of durable biofilms. The exopolysaccharides Pel, Psl and alginate essentially contribute to the biofilm matrix, but their secretion mechanisms are barely understood. Here, we reveal the architecture of the outer membrane complex PelBC for Pel export, where the essential periplasmic ring of twelve lipoproteins PelC is mounted on top of the nanodisc-embedded β-barrel PelB. The PelC assembly is stabilized by electrostatic contacts with the periplasmic rim of PelB and via the membrane-anchored acyl chains. The negatively charged interior of the PelB β-barrel forms a route for the cationic Pel exopolysaccharide. The β-barrel is sealed at the extracellular side, but molecular dynamic simulations suggest that the short loop Plug-S is sufficiently flexible to open a tunnel for the exopolysaccharide transport. This gating model is corroborated by single-channel conductivity measurements, where a deletion of Plug-S renders a constitutively open β-barrel. Our structural and functional analysis offers a comprehensive view on this pathogenicity-relevant complex and suggests the route taken by the exopolysaccharide at the final secretion step.
- Synthetic Membrane Systems, Institute of Biochemistry, Heinrich Heine University Düsseldorf, Düsseldorf, Germany.
Organizational Affiliation: