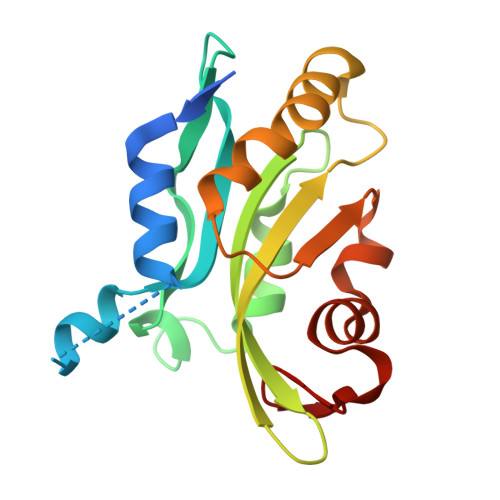
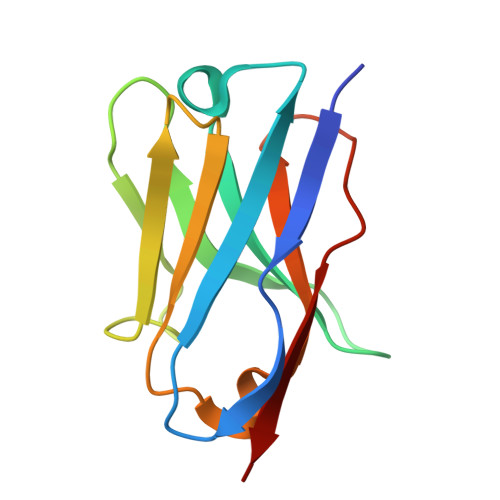
A high affinity Sybody blocks Cofilin-1 binding to F-actin in vitro and in cancer cells.
Paraschiakos, T., Li, J., Scholz, J., Han, S.J., Deckers, M., Pogenberg, V., Faix, J., Windhorst, S.(2025) Biochem Pharmacol 236: 116866-116866
- PubMed: 40064451
- DOI: https://doi.org/10.1016/j.bcp.2025.116866
- Primary Citation of Related Structures:
9H1F - PubMed Abstract:
Upregulation of the actin-severing protein Cofilin-1 is implicated in enhancing malignancy of various cancer types by promoting actin turnover and increasing cellular motility. Despite the importance of targeting Cofilin-1, currently there is a lack of inhibitors specifically targeting its actin-severing activity. To address this issue, we generated synthetic anti-Cofilin-1 nanobodies (Sybodies) that interfere with human Cofilin-1 binding to filamentous actin. We identified four high affinity Sybodies against human Cofilin-1 with dissociation constants (K D ) in the nanomolar range that inhibited G-actin sequestration, and actin-severing activity of Cofilin-1 in vitro. Notably, Sybody B12, with the lowest K D of approximately 27 nM, competitively blocked actin binding to Cofilin-1, and also inhibited G-actin sequestration of murine Cofilin-1. The crystal structure of the Sybody-B12-Cofilin-1 complex, resolved at 1.8 Å, revealed that Sybody B12 binds to the G-actin binding site of Cofilin-1, showing that Sybody B12 engages the same binding site on Cofilin-1 as actin. Consistently, transient expression of mPlum-tagged Sybody B12 in human H1299 lung cancer cells inhibited the formation of enhanced green fluorescent protein (EGFP)-Cofilin-actin rods. Notably, stable expression of Sybody B12 did not affect viability of H1299 cells, and no compensatory up-regulation of Cofilin-2 or actin-depolymerization factor (ADF) mRNA were detectable in Sybody B12 expressing H1299 cells. Together, these findings suggest that Sybody B12 exhibits a strong potential as tool for inhibiting the interaction of Cofilin-1 with actin. In addition, it could serve as a promising lead structure for designing Cofilin-1 inhibitors in silico.
- Department of Biochemistry and Signal Transduction, University Medical Center Hamburg-Eppendorf, Martinistrasse 52, 20246 Hamburg, Germany.
Organizational Affiliation: