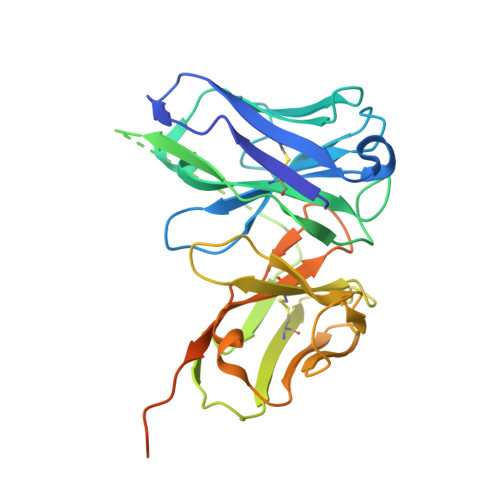
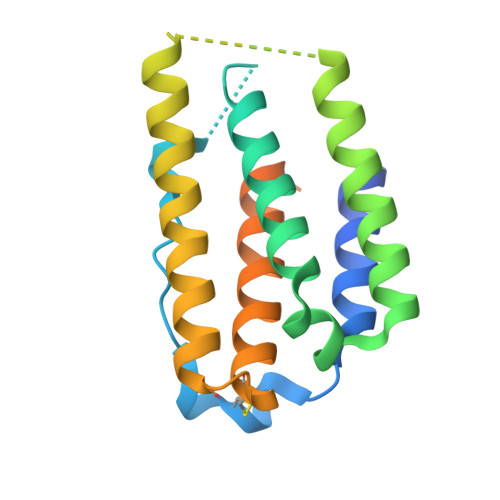
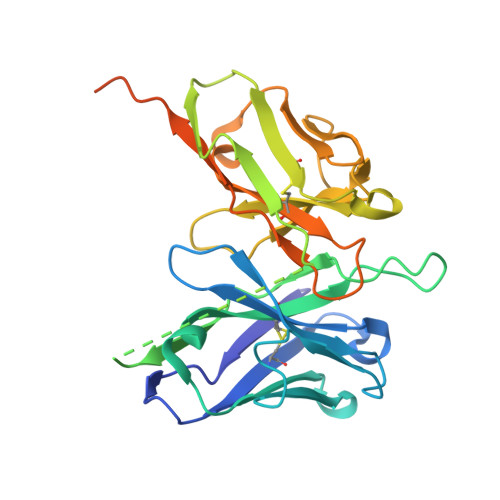
Temporal and structural insights into type-I interferons autoantibodies in severe COVID-19
Vanderkerken, M., Fournier, M., Dorgham, K., Bastard, P., Ahouzi, O., Duquerroy, S., Nguyen, N.K., Broutin, M., Charlet, M., Vandenberghe, A., Bizien, L., Da Mata-Jardin, O., Haouz, A., Belmondo, T., Hue, S., Borghesi, A., Rodriguez-Gallego, C., Vinh, D., Andreakos, V., Haerynck, F., Halwani, R., Hammerstrom, T.Q.P., Bjorkstrom, N., Strunz, B., Mogensen, T., Trouillet, S., Neven, B., Rodriguez-Gallego, C., Levy, R., Le Voyer, T., Delmonte, O., O'Farrelly, C., Riviere, J., Amador Borrero, B., Servettaz, A., Crickx, E., Michel, M., Puel, A., Abel, L., Luyt, C.E., Mathian, A., Amoura, Z., weill, J.C., Casanova, J.L., Rey, F.A., Gorochov, G., Chappert, P., Mahevas, M.To be published.