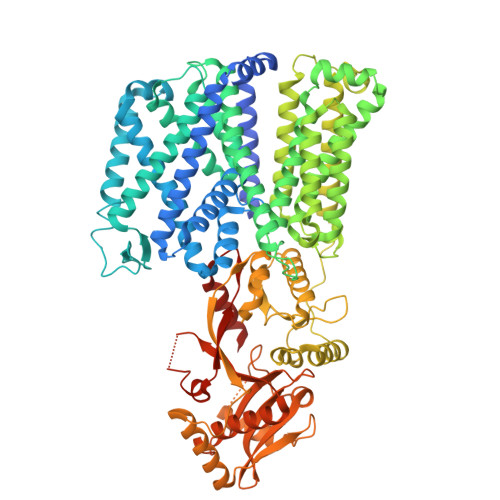
MprF from Pseudomonas aeruginosa is a promiscuous lipid scramblase with broad substrate specificity.
Hankins, M.T.K., Parrag, M., Garaeva, A.A., Earp, J.C., Seeger, M.A., Stansfeld, P.J., Bublitz, M.(2025) Sci Adv 11: eads9135-eads9135
- PubMed: 40203087
- DOI: https://doi.org/10.1126/sciadv.ads9135
- Primary Citation of Related Structures:
9GOE - PubMed Abstract:
The multiple peptide resistance factor (MprF) is a bifunctional membrane protein found in many bacteria, including Pseudomonas aeruginosa and Staphylococcus aureus . MprF modifies inner leaflet lipid headgroups through aminoacylation and translocates modified lipid to the outer leaflet. This activity provides increased resistance to antimicrobial agents. MprF presents a promising target in multiresistant pathogens, but structural information is limited and both substrate specificity and energization of MprF-mediated lipid transport are poorly understood. Here, we present the cryo-EM structure of MprF from P. aeruginosa ( Pa MprF) bound to a synthetic nanobody. Pa MprF adopts an "open" conformation with a wide, lipid-exposed groove on the periplasmic side that induces a local membrane deformation in molecular dynamics simulations. Using an in vitro liposome transport assay, we demonstrate that Pa MprF translocates a wide range of different lipids without an external energy source. This suggests that Pa MprF is the first dedicated lipid scramblase to be characterized in bacteria.
- Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, UK.
Organizational Affiliation: