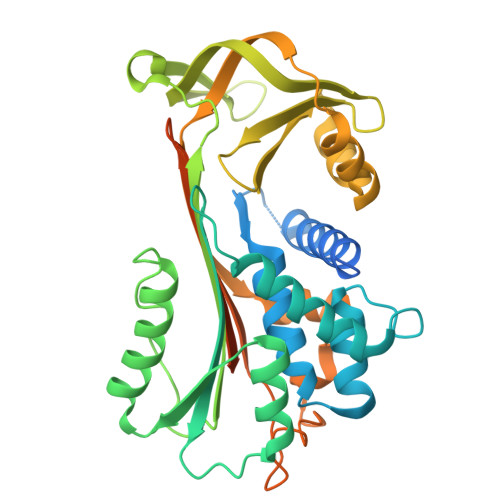
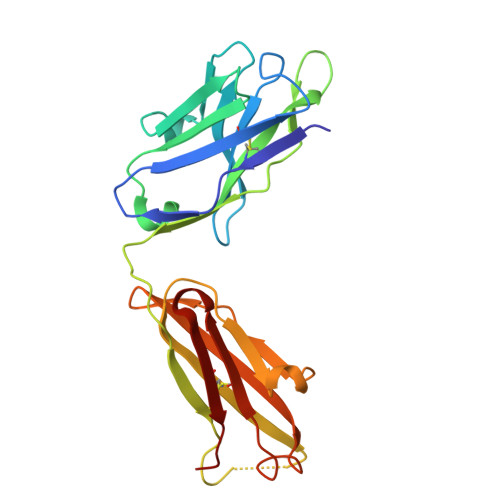
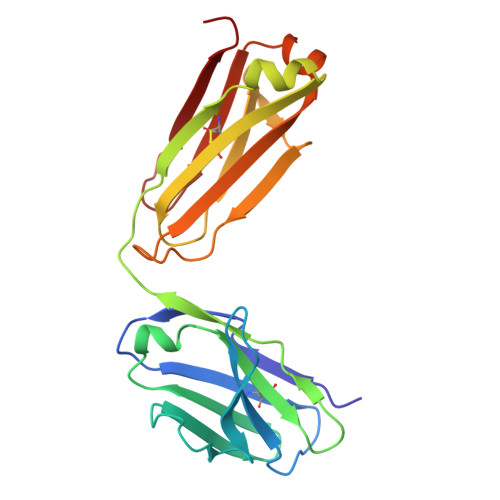
High-resolution characterization of ex vivo AAT polymers by solution-state NMR spectroscopy.
Lowen, S.M., Waudby, C.A., Jagger, A.M., Aldobiyan, I., Laffranchi, M., Fra, A., Christodoulou, J., Irving, J.A., Lomas, D.A.(2025) Sci Adv 11: eadu7064-eadu7064
- PubMed: 40333971
- DOI: https://doi.org/10.1126/sciadv.adu7064
- Primary Citation of Related Structures:
9GGP - PubMed Abstract:
Serpins, protease inhibitors whose regulated conformational instability renders them susceptible to mutations that cause misfolding, represent a system for the study of non-amyloid protein aggregation. The E342K "Z" variant of α-1-antitrypsin (AAT) undergoes oligomeric self-assembly into polymer chains that are associated with liver and lung pathologies in AAT deficiency. Structural characterization of polymers from human tissue has been limited by their heterogeneity and flexibility; here, we have studied their internal structure, which provides insights into the molecular linkage and the pathway by which they are formed. NMR spectra of heat-induced 13 C-ILV-methyl-labeled polymers, and 1 H-methyl spectra of liver-derived polymers, show equivalence to that of AAT in a post-protease-encounter conformation. This is corroborated by x-ray crystallography, which reveals a cryptic epitope recognized by the conformationally selective 2C1 antibody, common to both forms. These data definitively preclude most models of polymerization and are compatible with sequential intermolecular donation of the carboxyl terminus of one molecule into the next during polymer formation.
- UCL Respiratory, Rayne Institute, and the Institute of Structural and Molecular Biology, University College London, London WC1E 6JF, UK.
Organizational Affiliation: