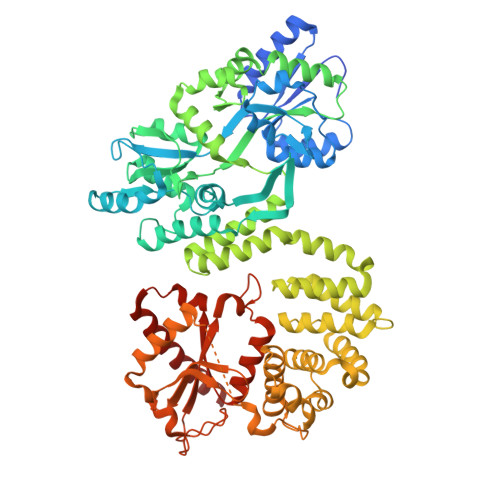
The Vsr-like protein FASTKD4 regulates the stability and polyadenylation of the MT-ND3 mRNA.
Yang, X., Stentenbach, M., Hughes, L.A., Siira, S.J., Lau, K., Hothorn, M., Martinou, J.C., Rackham, O., Filipovska, A.(2025) Nucleic Acids Res 53
- PubMed: 39727163
- DOI: https://doi.org/10.1093/nar/gkae1261
- Primary Citation of Related Structures:
9GEK - PubMed Abstract:
Expression of the compact mitochondrial genome is regulated by nuclear encoded, mitochondrially localized RNA-binding proteins (RBPs). RBPs regulate the lifecycles of mitochondrial RNAs from transcription to degradation by mediating RNA processing, maturation, stability and translation. The Fas-activated serine/threonine kinase (FASTK) family of RBPs has been shown to regulate and fine-tune discrete aspects of mitochondrial gene expression. Although the roles of specific targets of FASTK proteins have been elucidated, the molecular mechanisms of FASTK proteins in mitochondrial RNA metabolism remain unclear. Therefore, we resolved the structure of FASTKD4 at atomic level that includes the RAP domain and the two FAST motifs, creating a positively charged cavity resembling that of the very short patch repair endonuclease. Our biochemical studies show that FASTKD4 binds the canonical poly(A) tail of MT-ND3 enabling its maturation and translation. The in vitro role of FASTKD4 is consistent with its loss in cells that results in decreased MT-ND3 polyadenylation, which destabilizes this messenger RNA in mitochondria.
- Department of Molecular Cell Biology, University of Geneva, Quai Ernest-Ansermet 30, 1211 Geneva, Switzerland.
Organizational Affiliation: